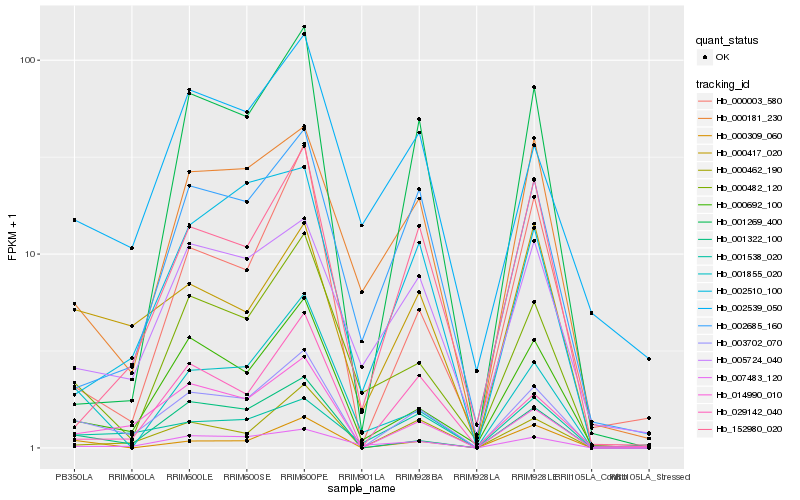
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003702_070 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_014990_010 |
0.1349808295 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v100031791mg, partial [Citrus clementina] |
| 3 |
Hb_000692_100 |
0.1508787919 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000462_190 |
0.1633271224 |
- |
- |
PREDICTED: thaumatin-like protein 1 [Jatropha curcas] |
| 5 |
Hb_001322_100 |
0.1659142241 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase [Vitis vinifera] |
| 6 |
Hb_000417_020 |
0.1740463948 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 7 |
Hb_000482_120 |
0.1759179748 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 8 |
Hb_007483_120 |
0.1790008587 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 9 |
Hb_001538_020 |
0.1835913971 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 10 |
Hb_001855_020 |
0.18426954 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_005724_040 |
0.187197322 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 12 |
Hb_000181_230 |
0.1884859859 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 13 |
Hb_029142_040 |
0.1915859326 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 14 |
Hb_152980_020 |
0.1943590711 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 15 |
Hb_002685_160 |
0.1959752415 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 16 |
Hb_001269_400 |
0.196237182 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 17 |
Hb_002510_100 |
0.1969629385 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 18 |
Hb_000309_060 |
0.1983268484 |
- |
- |
hypothetical protein JCGZ_09609 [Jatropha curcas] |
| 19 |
Hb_000003_580 |
0.1994643962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002539_050 |
0.1996480145 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |