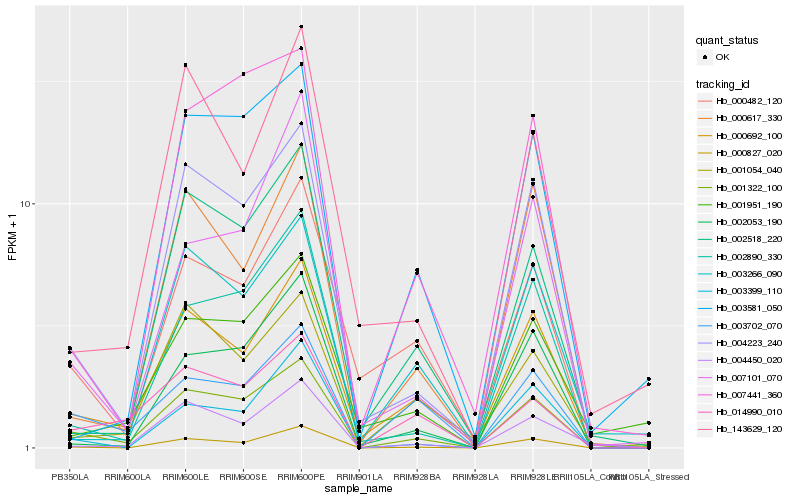
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001322_100 |
0.0 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase [Vitis vinifera] |
| 2 |
Hb_004223_240 |
0.1268831499 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 3 |
Hb_000692_100 |
0.1401334281 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_001951_190 |
0.15967804 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 5 |
Hb_002890_330 |
0.1635347134 |
- |
- |
caspase, putative [Ricinus communis] |
| 6 |
Hb_000827_020 |
0.1639406575 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_003702_070 |
0.1659142241 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_007441_360 |
0.1667375049 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 9 |
Hb_003581_050 |
0.1693732083 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 10 |
Hb_014990_010 |
0.1718756567 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v100031791mg, partial [Citrus clementina] |
| 11 |
Hb_003266_090 |
0.1724534128 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 12 |
Hb_002518_220 |
0.1743090348 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000617_330 |
0.1755850105 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 14 |
Hb_143629_120 |
0.1760696885 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 15 |
Hb_001054_040 |
0.1775051778 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 16 |
Hb_004450_020 |
0.1793050735 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 17 |
Hb_002053_190 |
0.1799729278 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003399_110 |
0.1804044561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000482_120 |
0.1821796193 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 20 |
Hb_007101_070 |
0.1852497318 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |