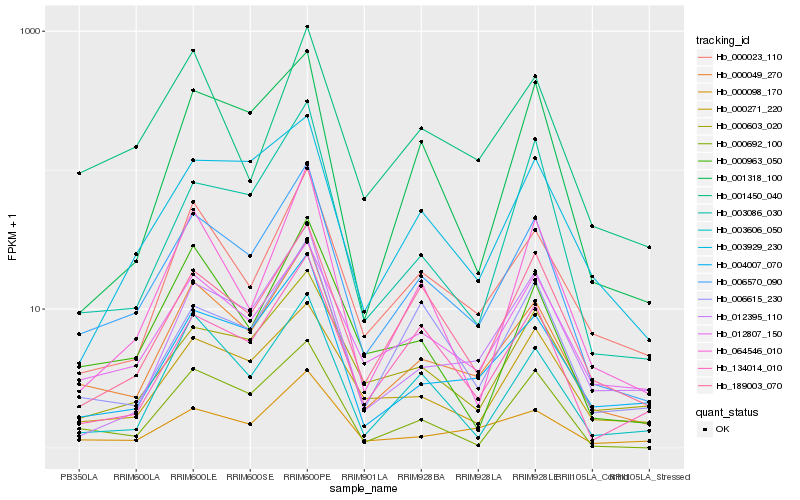
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006570_090 |
0.0 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 2 |
Hb_000049_270 |
0.0675284424 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012807_150 |
0.1174940599 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 4 |
Hb_004007_070 |
0.1212774721 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 5 |
Hb_134014_010 |
0.1252258439 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 6 |
Hb_000603_020 |
0.128573754 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000023_110 |
0.1285969285 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 8 |
Hb_000098_170 |
0.1289820786 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 9 |
Hb_000692_100 |
0.1296798634 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 10 |
Hb_189003_070 |
0.1313496052 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 11 |
Hb_003929_230 |
0.1364231674 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 12 |
Hb_006615_230 |
0.1364253885 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 13 |
Hb_000271_220 |
0.1364256556 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 14 |
Hb_003086_030 |
0.1374180011 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 15 |
Hb_000963_050 |
0.1405560473 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 16 |
Hb_064546_010 |
0.1412441692 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 17 |
Hb_003606_050 |
0.1420553413 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 18 |
Hb_001318_100 |
0.1444321659 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 19 |
Hb_012395_110 |
0.1467794652 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 20 |
Hb_001450_040 |
0.1471728643 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |