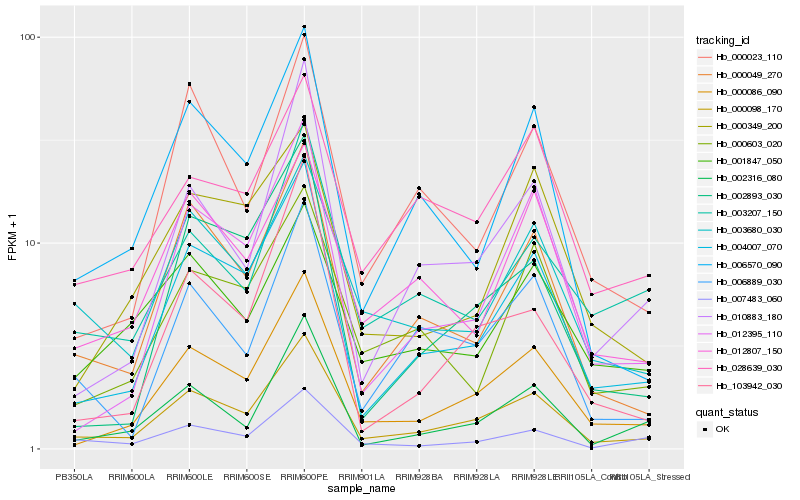
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_170 |
0.0 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 2 |
Hb_004007_070 |
0.0894720284 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 3 |
Hb_000086_090 |
0.0897786284 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 4 |
Hb_000049_270 |
0.1060672964 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006570_090 |
0.1289820786 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 6 |
Hb_006889_030 |
0.1309218527 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 7 |
Hb_012395_110 |
0.1316513913 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 8 |
Hb_003680_030 |
0.1325795505 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 9 |
Hb_002316_080 |
0.1347609808 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 10 |
Hb_010883_180 |
0.1361971142 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 11 |
Hb_012807_150 |
0.1414329012 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 12 |
Hb_000023_110 |
0.1430692445 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 13 |
Hb_007483_060 |
0.1442479576 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 14 |
Hb_028639_030 |
0.1465712683 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 15 |
Hb_000603_020 |
0.1486378241 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003207_150 |
0.1501176028 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 17 |
Hb_002893_030 |
0.1530917794 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 18 |
Hb_103942_030 |
0.1537289006 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000349_200 |
0.156586657 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g12500 [Jatropha curcas] |
| 20 |
Hb_001847_050 |
0.1571937849 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |