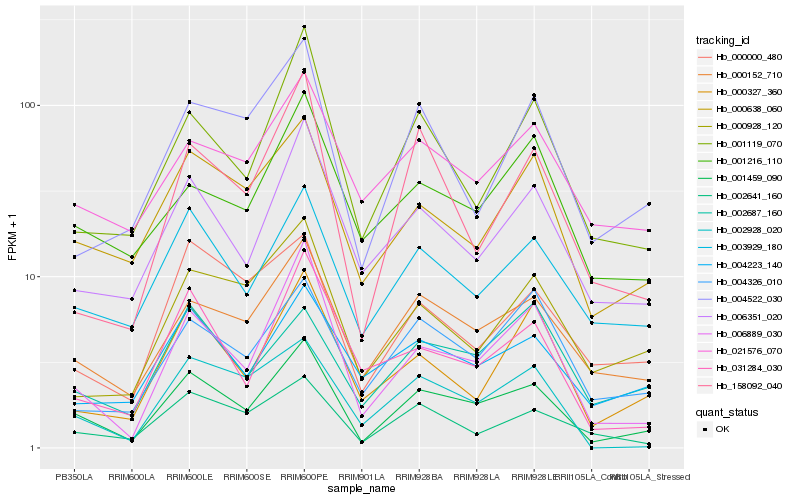
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001459_090 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_006889_030 |
0.1293251864 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000152_710 |
0.1357122647 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 4 |
Hb_003929_180 |
0.1482803947 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 5 |
Hb_000638_060 |
0.1494460973 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 6 |
Hb_006351_020 |
0.1524266152 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 7 |
Hb_002641_160 |
0.1610513319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001119_070 |
0.1612421739 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 9 |
Hb_000327_360 |
0.1694550315 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_002928_020 |
0.1718267644 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 11 |
Hb_004223_140 |
0.1719371818 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 12 |
Hb_004326_010 |
0.1729637239 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_158092_040 |
0.1731656592 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 14 |
Hb_031284_030 |
0.1733293349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004522_030 |
0.173681095 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 16 |
Hb_000000_480 |
0.1740509471 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001216_110 |
0.1755914502 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 18 |
Hb_021576_070 |
0.1763681875 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 19 |
Hb_002687_160 |
0.1763987395 |
- |
- |
PREDICTED: uncharacterized protein LOC105643395 [Jatropha curcas] |
| 20 |
Hb_000928_120 |
0.1764899717 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |