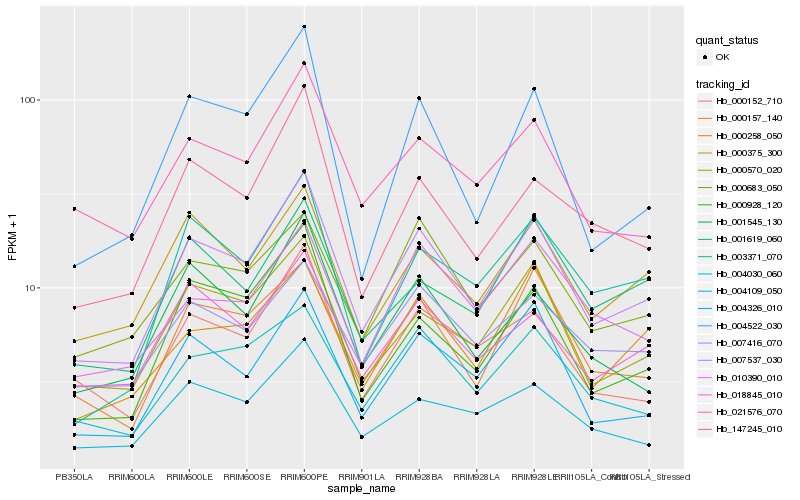
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007537_030 |
0.0 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000570_020 |
0.082154875 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 3 |
Hb_000157_140 |
0.0896445976 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_004030_060 |
0.0954391493 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 5 |
Hb_018845_010 |
0.096214617 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 6 |
Hb_021576_070 |
0.0964126995 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 7 |
Hb_004326_010 |
0.0966612923 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_000152_710 |
0.0966988198 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 9 |
Hb_004109_050 |
0.097731602 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 10 |
Hb_000375_300 |
0.0982618042 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 11 |
Hb_007416_070 |
0.101312536 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 12 |
Hb_000928_120 |
0.102598142 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 13 |
Hb_147245_010 |
0.1026141209 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 14 |
Hb_000683_050 |
0.1038666082 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 15 |
Hb_001545_130 |
0.1045681091 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 16 |
Hb_004522_030 |
0.1061071673 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 17 |
Hb_000258_050 |
0.1089892595 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003371_070 |
0.109123103 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_010390_010 |
0.1129038534 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 20 |
Hb_001619_060 |
0.1147259887 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |