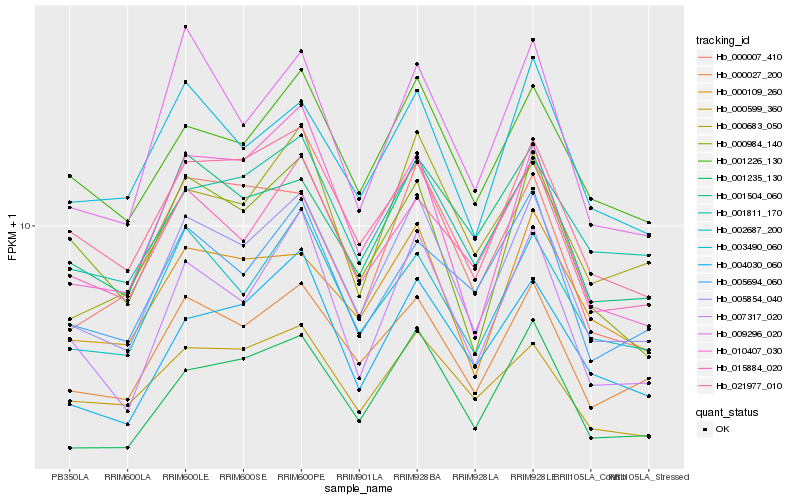
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005854_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000027_200 |
0.0559610771 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 3 |
Hb_007317_020 |
0.0798957201 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002687_200 |
0.0807845102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_021977_010 |
0.0823087485 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 6 |
Hb_001226_130 |
0.0832170672 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 7 |
Hb_015884_020 |
0.084338693 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 8 |
Hb_001235_130 |
0.0875574779 |
- |
- |
- |
| 9 |
Hb_003490_060 |
0.088266639 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 10 |
Hb_001504_060 |
0.0887240913 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 11 |
Hb_009296_020 |
0.0890901291 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000599_360 |
0.0901187039 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 13 |
Hb_000683_050 |
0.0914540573 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 14 |
Hb_010407_030 |
0.0919402632 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 15 |
Hb_001811_170 |
0.0923331834 |
- |
- |
dynamin, putative [Ricinus communis] |
| 16 |
Hb_004030_060 |
0.0926129993 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 17 |
Hb_000109_260 |
0.0926260591 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000984_140 |
0.0950093894 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 19 |
Hb_005694_060 |
0.0951911844 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 20 |
Hb_000007_410 |
0.0956349316 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |