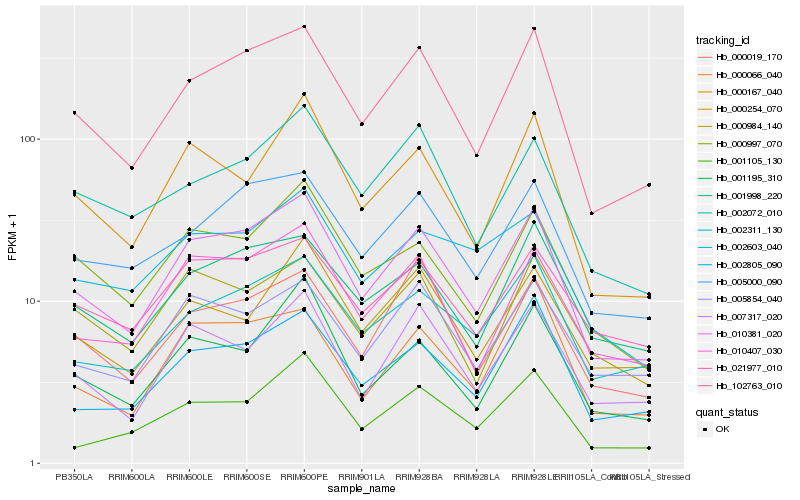
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010381_020 |
0.0 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 2 |
Hb_102763_010 |
0.0489470571 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 3 |
Hb_010407_030 |
0.0818877727 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 4 |
Hb_005000_090 |
0.0890099276 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 5 |
Hb_000167_040 |
0.0960812522 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 6 |
Hb_000997_070 |
0.0963168574 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 7 |
Hb_002311_130 |
0.0963541445 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001998_220 |
0.096593209 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002805_090 |
0.098065743 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 10 |
Hb_001195_310 |
0.1007651222 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_007317_020 |
0.1009331611 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_021977_010 |
0.1039484784 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 13 |
Hb_005854_040 |
0.1045251604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002072_010 |
0.1061630129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001105_130 |
0.107485417 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 16 |
Hb_000984_140 |
0.1107051701 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 17 |
Hb_000019_170 |
0.1110989519 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 18 |
Hb_000066_040 |
0.1129295552 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 19 |
Hb_000254_070 |
0.113196835 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 20 |
Hb_002603_040 |
0.1135800191 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2-like [Jatropha curcas] |