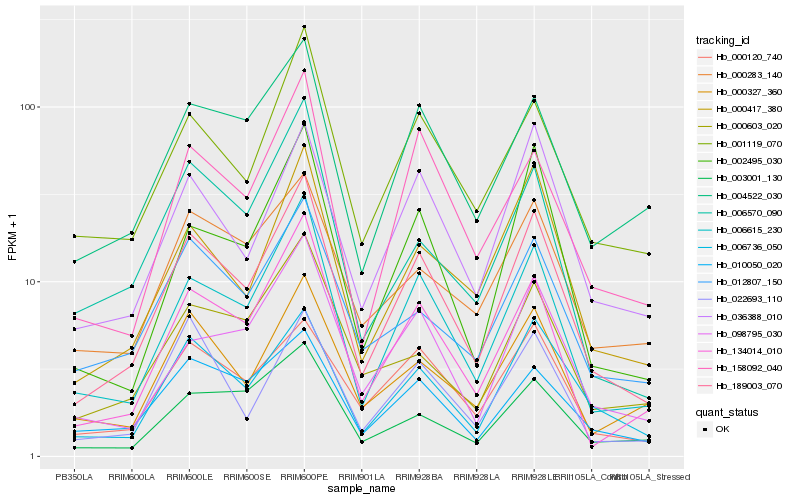
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_189003_070 |
0.0 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 2 |
Hb_006615_230 |
0.0776442542 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 3 |
Hb_006736_050 |
0.1000208675 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 4 |
Hb_134014_010 |
0.100842363 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 5 |
Hb_036388_010 |
0.1101449894 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 6 |
Hb_000417_380 |
0.1102735225 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |
| 7 |
Hb_001119_070 |
0.115379421 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 8 |
Hb_002495_030 |
0.1185619549 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000120_740 |
0.1228949239 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004522_030 |
0.1241188455 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 11 |
Hb_012807_150 |
0.1243321797 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 12 |
Hb_000327_360 |
0.124970371 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_158092_040 |
0.1257436298 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 14 |
Hb_010050_020 |
0.1260396351 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 15 |
Hb_098795_030 |
0.1268420843 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 16 |
Hb_006570_090 |
0.1313496052 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 17 |
Hb_000603_020 |
0.1317222753 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003001_130 |
0.1331856033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000283_140 |
0.1333338486 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_022693_110 |
0.135985775 |
- |
- |
phosphate transporter [Manihot esculenta] |