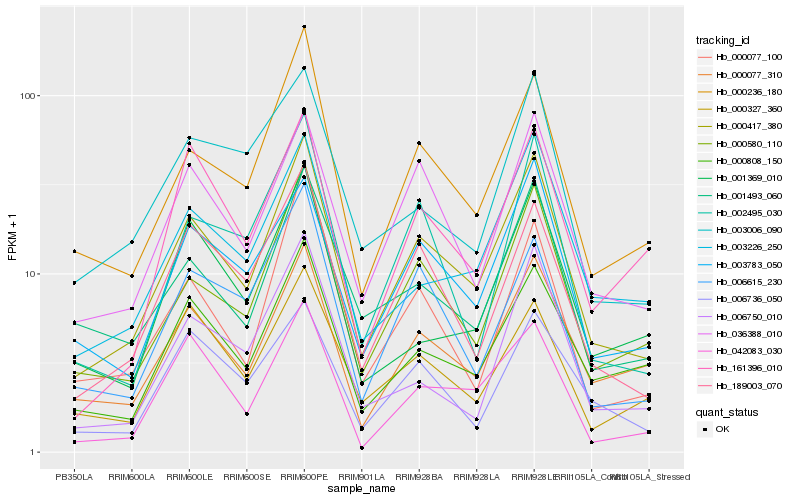
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_380 |
0.0 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |
| 2 |
Hb_000580_110 |
0.0902681201 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 3 |
Hb_000236_180 |
0.1001571834 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 4 |
Hb_036388_010 |
0.1059035519 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 5 |
Hb_189003_070 |
0.1102735225 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 6 |
Hb_000077_310 |
0.1111561179 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 7 |
Hb_003783_050 |
0.1156942812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001493_060 |
0.1204170124 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 9 |
Hb_003226_250 |
0.1212178061 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 10 |
Hb_000808_150 |
0.1214477668 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 11 |
Hb_002495_030 |
0.1272177675 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 12 |
Hb_042083_030 |
0.1278688604 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_006615_230 |
0.1324660197 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 14 |
Hb_000327_360 |
0.1378249472 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_006736_050 |
0.1379466834 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 16 |
Hb_003006_090 |
0.1396439755 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 17 |
Hb_000077_100 |
0.1396530365 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_006750_010 |
0.1403979351 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 19 |
Hb_161396_010 |
0.143222232 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 20 |
Hb_001369_010 |
0.1443158843 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |