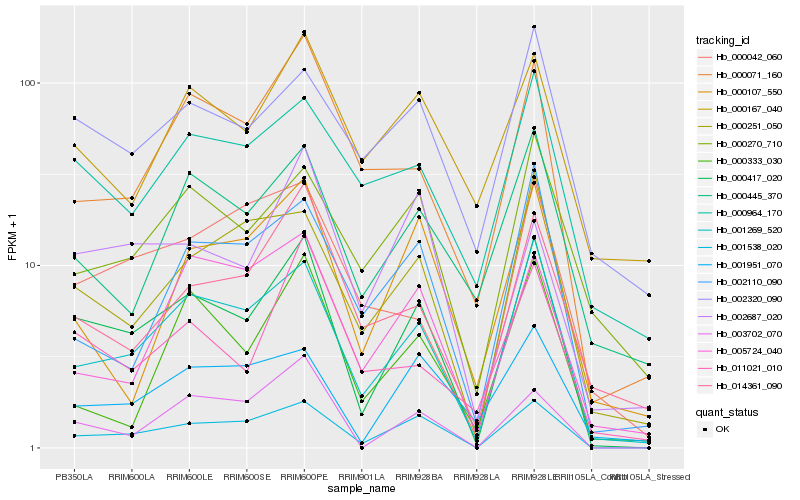
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 2 |
Hb_001538_020 |
0.1073511475 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 3 |
Hb_001269_520 |
0.1181809676 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 4 |
Hb_000251_050 |
0.173005865 |
- |
- |
hypothetical protein JCGZ_21678 [Jatropha curcas] |
| 5 |
Hb_003702_070 |
0.1740463948 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000071_160 |
0.1889520808 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 7 |
Hb_002320_090 |
0.1910688845 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 8 |
Hb_011021_010 |
0.1911078946 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_000270_710 |
0.1920962927 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_014361_090 |
0.1926194887 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 11 |
Hb_000964_170 |
0.1928647672 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 12 |
Hb_001951_070 |
0.1935331992 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 13 |
Hb_002110_090 |
0.1970088078 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 14 |
Hb_000445_370 |
0.1982905245 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 15 |
Hb_002687_020 |
0.2040846322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000042_060 |
0.2054394949 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 17 |
Hb_000107_550 |
0.2070173662 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 18 |
Hb_005724_040 |
0.2079519598 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 19 |
Hb_000167_040 |
0.2108489273 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 20 |
Hb_000333_030 |
0.2118372428 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |