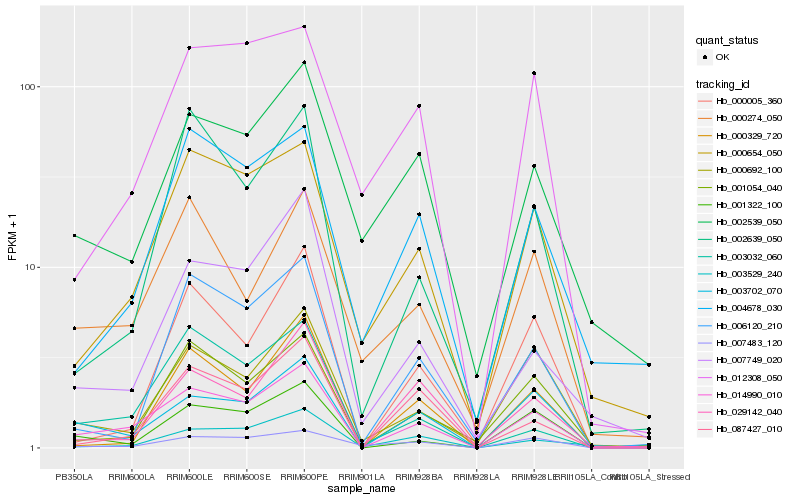
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014990_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v100031791mg, partial [Citrus clementina] |
| 2 |
Hb_003702_070 |
0.1349808295 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000654_050 |
0.1625442883 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 4 |
Hb_000692_100 |
0.1683893198 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 5 |
Hb_001322_100 |
0.1718756567 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase [Vitis vinifera] |
| 6 |
Hb_001054_040 |
0.1722662805 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 7 |
Hb_087427_010 |
0.1802161565 |
- |
- |
hypothetical protein PRUPE_ppa007748mg [Prunus persica] |
| 8 |
Hb_006120_210 |
0.1827661915 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 9 |
Hb_000274_050 |
0.1834988122 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 10 |
Hb_007483_120 |
0.1871567238 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 11 |
Hb_002639_050 |
0.1874593745 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 12 |
Hb_000329_720 |
0.1874860705 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 13 |
Hb_000005_360 |
0.1875689388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004678_030 |
0.1893731638 |
- |
- |
fructokinase [Manihot esculenta] |
| 15 |
Hb_029142_040 |
0.1896549365 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 16 |
Hb_002539_050 |
0.1900885551 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 17 |
Hb_003032_060 |
0.1907678664 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 18 |
Hb_007749_020 |
0.1911990602 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_012308_050 |
0.193620381 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 20 |
Hb_003529_240 |
0.1945706981 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7-like isoform X2 [Gossypium raimondii] |