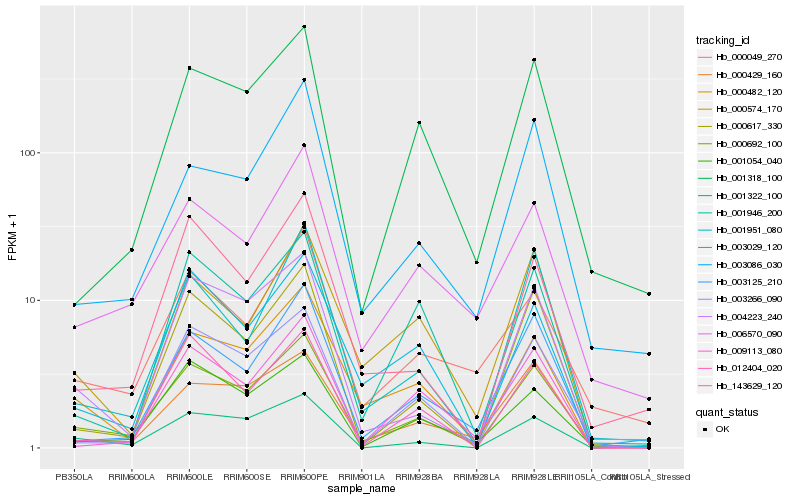
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000692_100 |
0.0 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000482_120 |
0.1216807299 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 3 |
Hb_004223_240 |
0.1248657314 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 4 |
Hb_001054_040 |
0.127975849 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 5 |
Hb_000617_330 |
0.1288152091 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 6 |
Hb_006570_090 |
0.1296798634 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 7 |
Hb_001318_100 |
0.1316392874 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 8 |
Hb_003029_120 |
0.135350436 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 9 |
Hb_000429_160 |
0.1358263812 |
- |
- |
PREDICTED: uncharacterized protein LOC105645177 [Jatropha curcas] |
| 10 |
Hb_003125_210 |
0.1360235441 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 11 |
Hb_001951_080 |
0.1374667275 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 12 |
Hb_003266_090 |
0.1391291994 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 13 |
Hb_001322_100 |
0.1401334281 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase [Vitis vinifera] |
| 14 |
Hb_009113_080 |
0.1426010133 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 15 |
Hb_012404_020 |
0.1426778649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001946_200 |
0.1432152144 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_143629_120 |
0.1443889139 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 18 |
Hb_003086_030 |
0.1452502114 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 19 |
Hb_000574_170 |
0.1469863235 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 20 |
Hb_000049_270 |
0.1473991661 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |