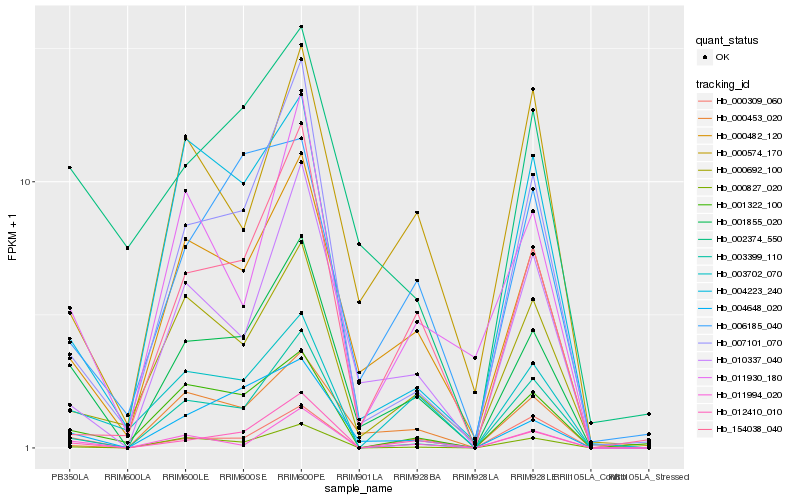
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001855_020 |
0.0 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000482_120 |
0.1253432236 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 3 |
Hb_004648_020 |
0.1499099526 |
- |
- |
PREDICTED: uncharacterized protein LOC105643062 [Jatropha curcas] |
| 4 |
Hb_012410_010 |
0.1700430573 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 5 |
Hb_010337_040 |
0.1727256678 |
- |
- |
- |
| 6 |
Hb_007101_070 |
0.1789361713 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 7 |
Hb_003702_070 |
0.18426954 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000692_100 |
0.1897501788 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000309_060 |
0.1917705249 |
- |
- |
hypothetical protein JCGZ_09609 [Jatropha curcas] |
| 10 |
Hb_001322_100 |
0.1929353498 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase [Vitis vinifera] |
| 11 |
Hb_011930_180 |
0.1961553002 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002374_550 |
0.1970943427 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 13 |
Hb_003399_110 |
0.198090082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000453_020 |
0.2017707594 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000827_020 |
0.2018099953 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_011994_020 |
0.2064883447 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 17 |
Hb_154038_040 |
0.2067147826 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 18 |
Hb_004223_240 |
0.208148044 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 19 |
Hb_006185_040 |
0.2096117724 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 20 |
Hb_000574_170 |
0.2111569245 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |