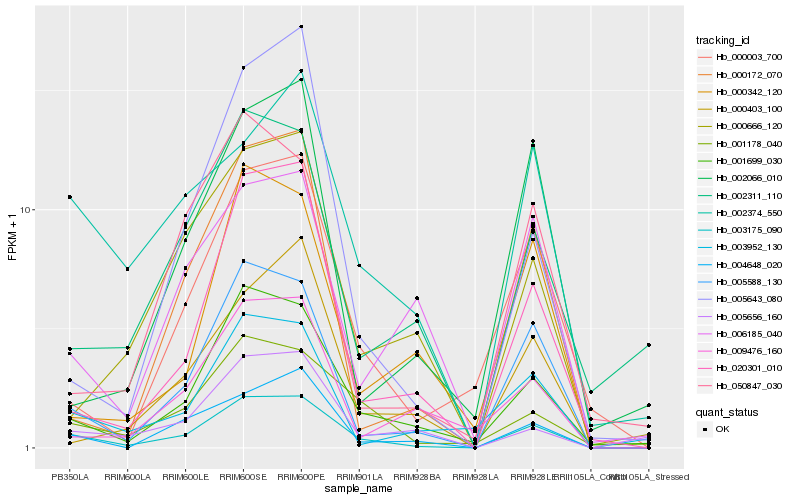
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003175_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 2 |
Hb_001699_030 |
0.1417805784 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005588_130 |
0.1460454467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001178_040 |
0.1695518421 |
desease resistance |
Gene Name: ABC_trans_N |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 5 |
Hb_004648_020 |
0.1695919714 |
- |
- |
PREDICTED: uncharacterized protein LOC105643062 [Jatropha curcas] |
| 6 |
Hb_020301_010 |
0.1890917267 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_002374_550 |
0.1915171938 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 8 |
Hb_000003_700 |
0.1937197027 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 9 |
Hb_003952_130 |
0.204882808 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 10 |
Hb_005656_160 |
0.210633655 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |
| 11 |
Hb_002311_110 |
0.2108790458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000172_070 |
0.2142332564 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_000666_120 |
0.2148738398 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_006185_040 |
0.2159784732 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 15 |
Hb_000403_100 |
0.2168294174 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 16 |
Hb_009476_160 |
0.2241943001 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 17 |
Hb_005643_080 |
0.2246385237 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 18 |
Hb_000342_120 |
0.2263492475 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 19 |
Hb_050847_030 |
0.2282647785 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 20 |
Hb_002066_010 |
0.2339902076 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |