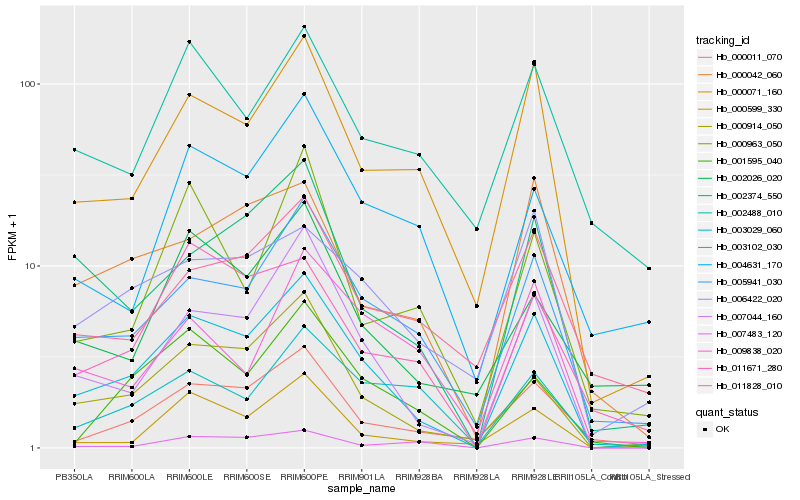
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003102_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644418 [Jatropha curcas] |
| 2 |
Hb_007044_160 |
0.121540783 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Populus euphratica] |
| 3 |
Hb_005941_030 |
0.1380239883 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000914_050 |
0.1415385184 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 5 |
Hb_000071_160 |
0.1443037743 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 6 |
Hb_000011_070 |
0.1622080098 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 7 |
Hb_002374_550 |
0.1657933686 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 8 |
Hb_000599_330 |
0.1729590245 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 9 |
Hb_000042_060 |
0.1814236766 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 10 |
Hb_001595_040 |
0.1847153271 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 11 |
Hb_011671_280 |
0.1863991848 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF106-like [Populus euphratica] |
| 12 |
Hb_006422_020 |
0.1868258988 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 13 |
Hb_003029_060 |
0.1937134901 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002026_020 |
0.2005159425 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 15 |
Hb_004631_170 |
0.2011159716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007483_120 |
0.2014230704 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 17 |
Hb_011828_010 |
0.2015415197 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 18 |
Hb_002488_010 |
0.2017704606 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 19 |
Hb_000963_050 |
0.2051169167 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 20 |
Hb_009838_020 |
0.2059427148 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |