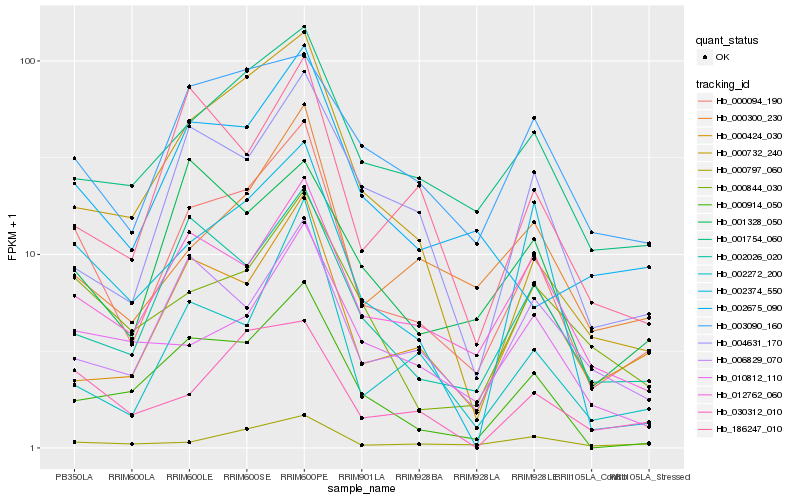
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_190 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 2 |
Hb_012762_060 |
0.158046222 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_001754_060 |
0.1634763874 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_002026_020 |
0.1668645623 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 5 |
Hb_000914_050 |
0.1705898631 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 6 |
Hb_000844_030 |
0.1744051965 |
- |
- |
ent-kaurene oxidase, putative [Ricinus communis] |
| 7 |
Hb_002374_550 |
0.1747100227 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 8 |
Hb_030312_010 |
0.1762938275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000300_230 |
0.1771818001 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 10 |
Hb_002675_090 |
0.1782100012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000732_240 |
0.1785605988 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 12 |
Hb_000797_060 |
0.1789469583 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 13 |
Hb_003090_160 |
0.184611113 |
- |
- |
- |
| 14 |
Hb_000424_030 |
0.1879487762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006829_070 |
0.189807735 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 16 |
Hb_186247_010 |
0.1899120265 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 17 |
Hb_002272_200 |
0.1907241918 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004631_170 |
0.1918152157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010812_110 |
0.1924933087 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 20 |
Hb_001328_050 |
0.195430792 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |