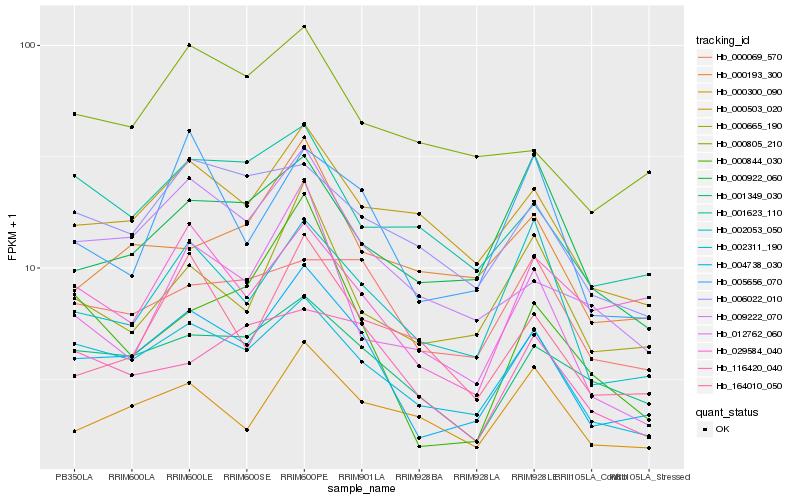
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004738_030 |
0.0 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 2 |
Hb_002311_190 |
0.1022021125 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 3 |
Hb_009222_070 |
0.131532749 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_001349_030 |
0.1333569467 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002053_050 |
0.1343124086 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 6 |
Hb_000503_020 |
0.1404570433 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 7 |
Hb_000665_190 |
0.1453067517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005656_070 |
0.1462032887 |
- |
- |
PREDICTED: cryptochrome DASH, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_000300_090 |
0.1507743517 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_006022_010 |
0.1535747672 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 11 |
Hb_164010_050 |
0.1548169451 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 12 |
Hb_000069_570 |
0.1557277836 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001623_110 |
0.1571602669 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_012762_060 |
0.1576515249 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_029584_040 |
0.1586474336 |
- |
- |
hypothetical protein Csa_1G586790 [Cucumis sativus] |
| 16 |
Hb_000193_300 |
0.159090598 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 17 |
Hb_116420_040 |
0.1595409797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000844_030 |
0.1606018024 |
- |
- |
ent-kaurene oxidase, putative [Ricinus communis] |
| 19 |
Hb_000922_060 |
0.1608606546 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 20 |
Hb_000805_210 |
0.1610443266 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |