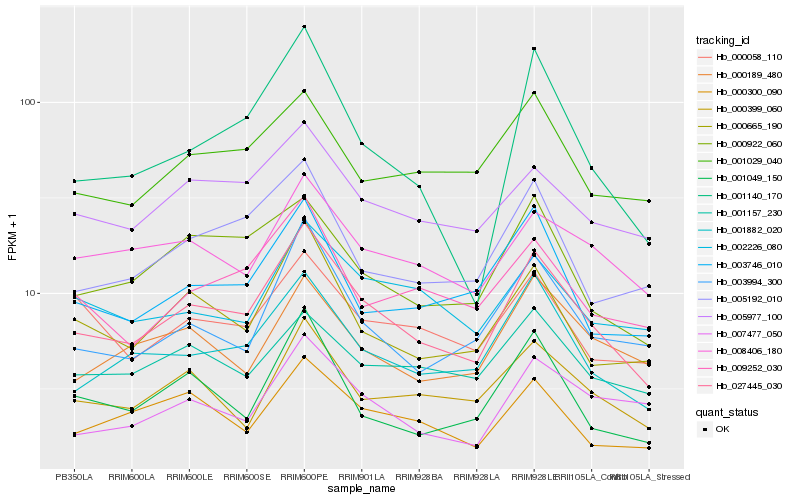
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027445_030 |
0.0 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_001882_020 |
0.0926467002 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 3 |
Hb_000665_190 |
0.1017496935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003994_300 |
0.1078553133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000922_060 |
0.1100365671 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 6 |
Hb_005977_100 |
0.1122508747 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 7 |
Hb_008406_180 |
0.1128031372 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_000399_060 |
0.1152750802 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 9 |
Hb_009252_030 |
0.115460057 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 10 |
Hb_005192_010 |
0.1160016832 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 11 |
Hb_000300_090 |
0.1160182887 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_003746_010 |
0.1161079479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002226_080 |
0.1205672814 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001029_040 |
0.1213875427 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 15 |
Hb_000058_110 |
0.1230145539 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001157_230 |
0.1238465055 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 17 |
Hb_001140_170 |
0.1241733408 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 18 |
Hb_000189_480 |
0.1245887648 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 19 |
Hb_007477_050 |
0.1253017298 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001049_150 |
0.1270627119 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |