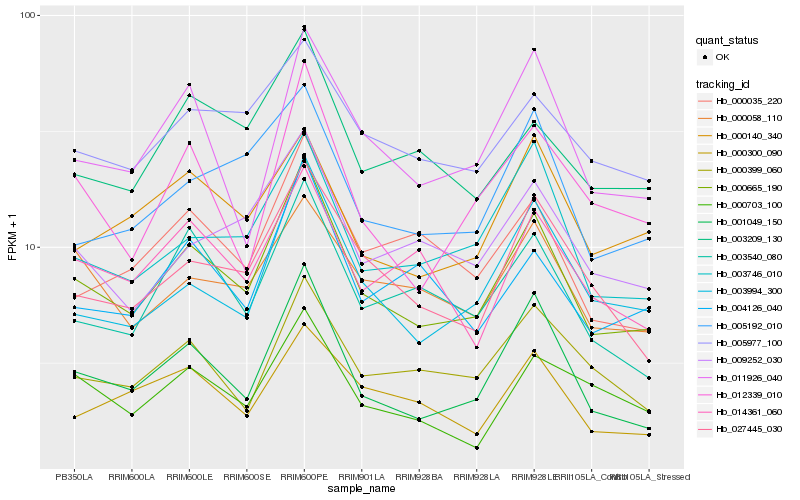
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000665_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001049_150 |
0.0805803649 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 3 |
Hb_003994_300 |
0.0997438272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_027445_030 |
0.1017496935 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_003746_010 |
0.1052364378 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003540_080 |
0.1053080079 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_011926_040 |
0.1099248968 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005192_010 |
0.1121930566 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 9 |
Hb_012339_010 |
0.1132548231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003209_130 |
0.1135508437 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 11 |
Hb_009252_030 |
0.1139596258 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 12 |
Hb_000035_220 |
0.1142224342 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 13 |
Hb_004126_040 |
0.1150073256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000399_060 |
0.1151118547 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 15 |
Hb_005977_100 |
0.1159772963 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 16 |
Hb_000058_110 |
0.1166811963 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 isoform X1 [Jatropha curcas] |
| 17 |
Hb_014361_060 |
0.1178480452 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 18 |
Hb_000300_090 |
0.1181846292 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_000140_340 |
0.1210956896 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 20 |
Hb_000703_100 |
0.1220710262 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase-like [Jatropha curcas] |