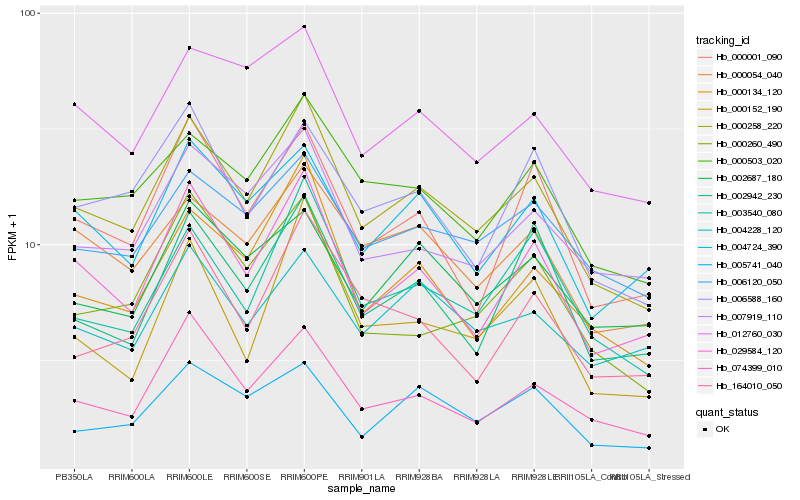
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_220 |
0.0 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_029584_120 |
0.0701744142 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000134_120 |
0.0804309018 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 4 |
Hb_000503_020 |
0.0824428107 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 5 |
Hb_074399_010 |
0.0826708256 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 6 |
Hb_000054_040 |
0.0890571957 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 7 |
Hb_003540_080 |
0.0900981025 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_007919_110 |
0.093049173 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 9 |
Hb_006120_050 |
0.0944733049 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 10 |
Hb_004724_390 |
0.0956009791 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 11 |
Hb_000001_090 |
0.0965668193 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 12 |
Hb_012760_030 |
0.098315701 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 13 |
Hb_006588_160 |
0.0987438667 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_005741_040 |
0.1008839209 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 15 |
Hb_002687_180 |
0.103732962 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 16 |
Hb_000260_490 |
0.1049346082 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 17 |
Hb_002942_230 |
0.1050377188 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 18 |
Hb_164010_050 |
0.1055535741 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 19 |
Hb_004228_120 |
0.106242827 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 20 |
Hb_000152_190 |
0.107507125 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |