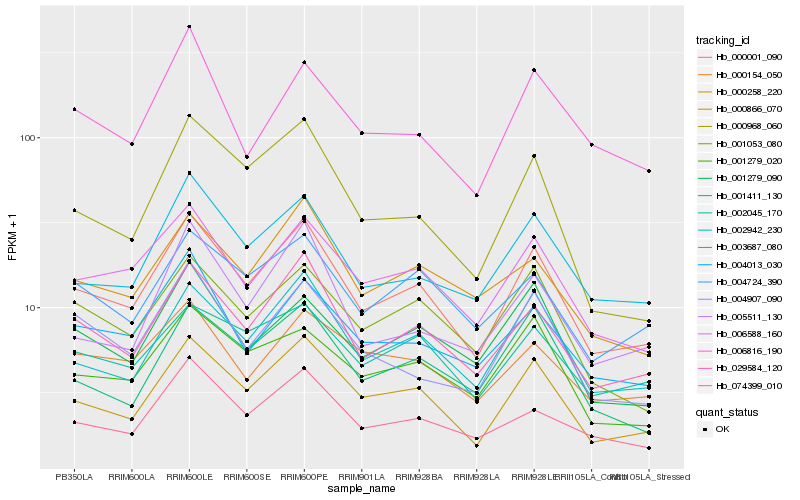
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000001_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 2 |
Hb_000866_070 |
0.0496613201 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 3 |
Hb_029584_120 |
0.0624192547 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006588_160 |
0.074002964 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_002942_230 |
0.0745080648 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 6 |
Hb_003687_080 |
0.0835049788 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 7 |
Hb_074399_010 |
0.0877388222 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 8 |
Hb_004013_030 |
0.0939891391 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 9 |
Hb_001279_020 |
0.0952046391 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 10 |
Hb_004724_390 |
0.0956057555 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 11 |
Hb_001053_080 |
0.096180149 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 12 |
Hb_002045_170 |
0.0962975964 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 13 |
Hb_000258_220 |
0.0965668193 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000968_060 |
0.0991739813 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 15 |
Hb_001279_090 |
0.1022642612 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 16 |
Hb_000154_050 |
0.1027782042 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 17 |
Hb_004907_090 |
0.1032448435 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_001411_130 |
0.103630219 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006816_190 |
0.105617467 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 20 |
Hb_005511_130 |
0.1079352293 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |