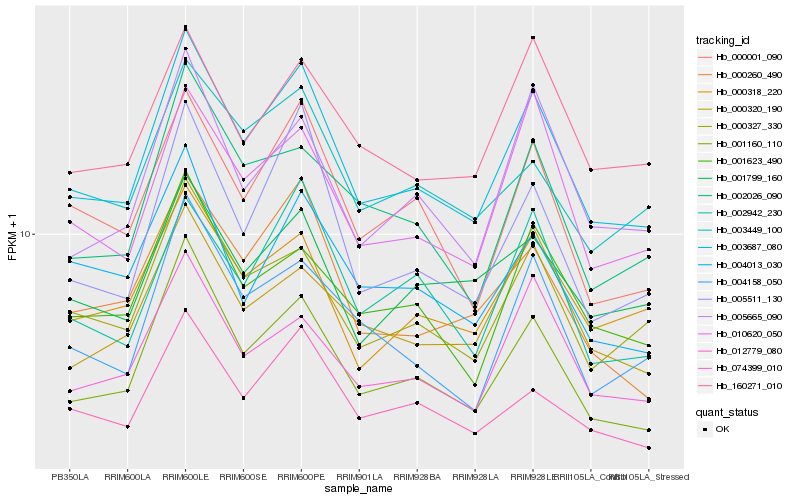
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003687_080 |
0.0 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 2 |
Hb_001160_110 |
0.0669290865 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 3 |
Hb_000327_330 |
0.0782592582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_074399_010 |
0.083182885 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 5 |
Hb_000001_090 |
0.0835049788 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 6 |
Hb_005511_130 |
0.0869436556 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 7 |
Hb_000318_220 |
0.0876578126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010620_050 |
0.0884336917 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 9 |
Hb_005665_090 |
0.0887041102 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 10 |
Hb_000260_490 |
0.0892479598 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 11 |
Hb_001623_490 |
0.0897941574 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 12 |
Hb_012779_080 |
0.0901866079 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 13 |
Hb_004158_050 |
0.0940401425 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 14 |
Hb_002026_090 |
0.0953861825 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 15 |
Hb_003449_100 |
0.0969979702 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 16 |
Hb_160271_010 |
0.097218103 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002942_230 |
0.0976226767 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 18 |
Hb_001799_160 |
0.0977707439 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_000320_190 |
0.098742856 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_004013_030 |
0.1007827056 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |