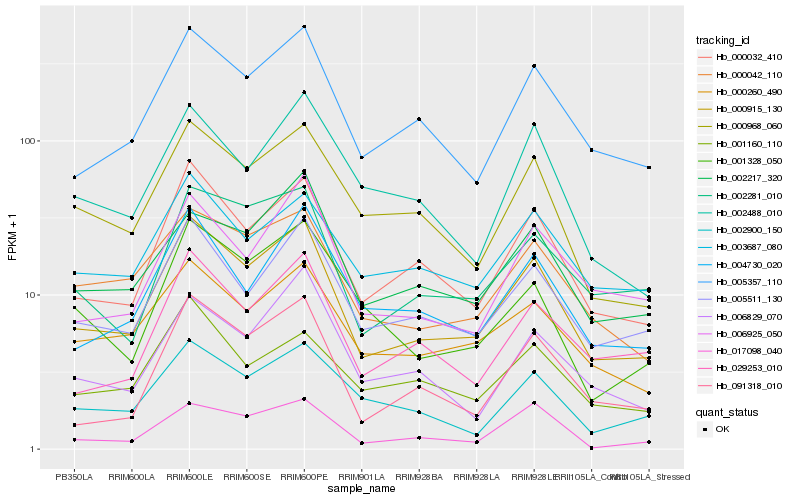
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000915_130 |
0.0 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 2 |
Hb_000032_410 |
0.0765624493 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 3 |
Hb_005357_110 |
0.0895034483 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 4 |
Hb_005511_130 |
0.09371733 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 5 |
Hb_000042_110 |
0.1063548059 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 6 |
Hb_000260_490 |
0.1081691282 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 7 |
Hb_004730_020 |
0.1117492705 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 8 |
Hb_000968_060 |
0.1117523231 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 9 |
Hb_002281_010 |
0.1132613401 |
- |
- |
hypothetical protein POPTR_0001s47550g [Populus trichocarpa] |
| 10 |
Hb_091318_010 |
0.1138284209 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 11 |
Hb_029253_010 |
0.1149233712 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 12 |
Hb_006925_050 |
0.1164792029 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 13 |
Hb_002217_320 |
0.1175686117 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002900_150 |
0.1197920675 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 15 |
Hb_003687_080 |
0.1217733096 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 16 |
Hb_001328_050 |
0.1282805678 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 17 |
Hb_001160_110 |
0.1287317154 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 18 |
Hb_017098_040 |
0.1307170125 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 19 |
Hb_006829_070 |
0.1307545177 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 20 |
Hb_002488_010 |
0.1313683188 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |