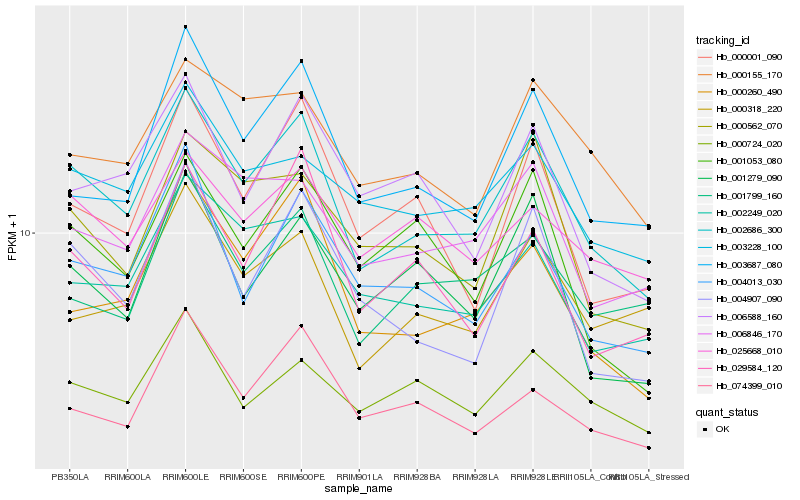
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_300 |
0.0 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 2 |
Hb_003687_080 |
0.1020541615 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 3 |
Hb_006846_170 |
0.1034674725 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 4 |
Hb_000260_490 |
0.1039310519 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 5 |
Hb_004013_030 |
0.1041975066 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 6 |
Hb_002249_020 |
0.1043409395 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 7 |
Hb_003228_100 |
0.1051368217 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 8 |
Hb_004907_090 |
0.10586663 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000155_170 |
0.1097240826 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |
| 10 |
Hb_000001_090 |
0.1101284287 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 11 |
Hb_000724_020 |
0.1104987104 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 12 |
Hb_000318_220 |
0.1111435905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001799_160 |
0.1112096159 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_000562_070 |
0.1123109869 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006588_160 |
0.1136772011 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_025668_010 |
0.1140637893 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_001279_090 |
0.1147330477 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 18 |
Hb_074399_010 |
0.1156646606 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 19 |
Hb_029584_120 |
0.1157727618 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001053_080 |
0.1160261596 |
- |
- |
OsCesA3 protein [Morus notabilis] |