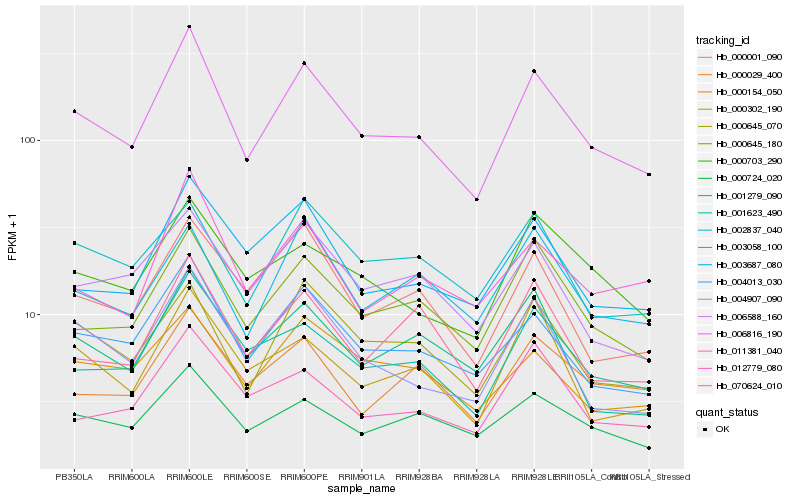
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_190 |
0.0 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 2 |
Hb_004013_030 |
0.0767801856 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 3 |
Hb_001623_490 |
0.0910344599 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000645_180 |
0.0945795468 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000645_070 |
0.0971861639 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_004907_090 |
0.0999113974 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_003687_080 |
0.1055100155 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 8 |
Hb_000001_090 |
0.105617467 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 9 |
Hb_011381_040 |
0.1075911864 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000724_020 |
0.1082121265 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 11 |
Hb_000703_290 |
0.1093594107 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 12 |
Hb_000154_050 |
0.1095971964 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 13 |
Hb_006588_160 |
0.1100181223 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_012779_080 |
0.1133255353 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 15 |
Hb_003058_100 |
0.1142514494 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000029_400 |
0.1152030808 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002837_040 |
0.1152737549 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 18 |
Hb_070624_010 |
0.1154992061 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 19 |
Hb_001279_090 |
0.115719612 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 20 |
Hb_000302_190 |
0.1167970424 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |