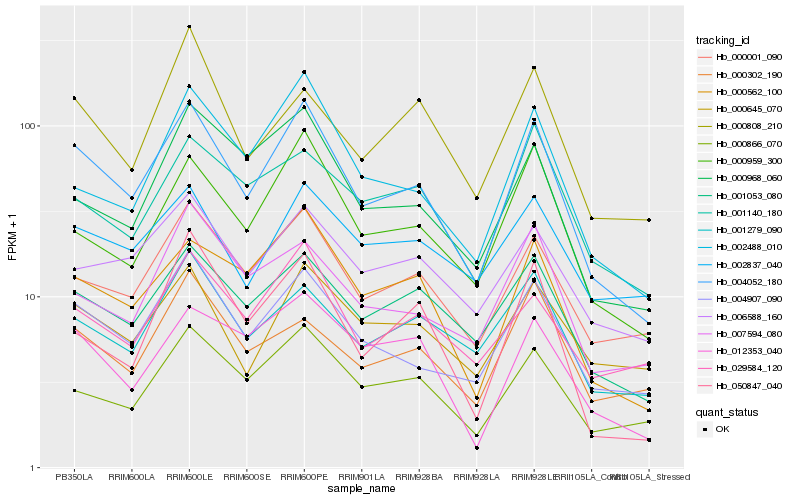
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004052_180 |
0.0 |
- |
- |
PREDICTED: protein COBRA-like [Jatropha curcas] |
| 2 |
Hb_004907_090 |
0.0963430435 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000562_100 |
0.1085099216 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 4 |
Hb_000001_090 |
0.112261002 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 5 |
Hb_001053_080 |
0.1132429727 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 6 |
Hb_000959_300 |
0.1155299152 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 7 |
Hb_002488_010 |
0.1205604755 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 8 |
Hb_000866_070 |
0.1225438377 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 9 |
Hb_000968_060 |
0.1292569673 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 10 |
Hb_029584_120 |
0.1304789682 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_012353_040 |
0.1318607526 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_006588_160 |
0.1318738276 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000645_070 |
0.1321557453 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_001279_090 |
0.1322053083 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 15 |
Hb_050847_040 |
0.1360145706 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 16 |
Hb_007594_080 |
0.1389164765 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 17 |
Hb_001140_180 |
0.1417360233 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 18 |
Hb_002837_040 |
0.1421986267 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 19 |
Hb_000808_210 |
0.1438582099 |
- |
- |
hypothetical protein EUTSA_v10023481mg [Eutrema salsugineum] |
| 20 |
Hb_000302_190 |
0.1459441244 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |