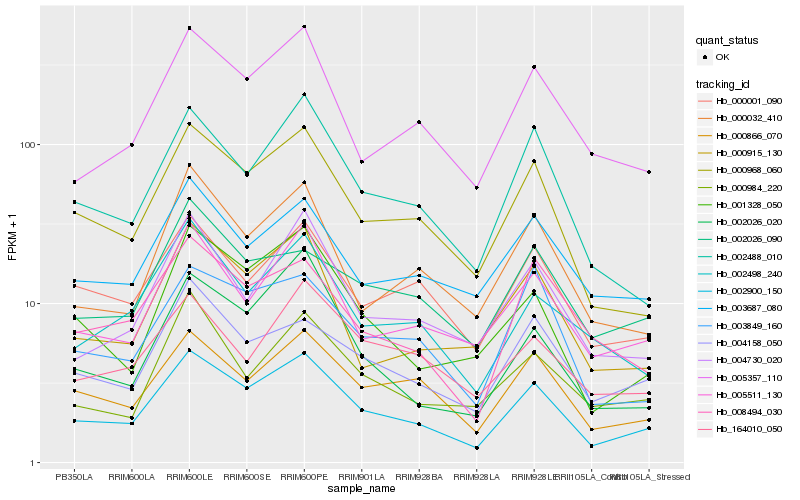
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002900_150 |
0.0 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 2 |
Hb_000968_060 |
0.0881828228 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 3 |
Hb_000866_070 |
0.1028785509 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 4 |
Hb_002488_010 |
0.1071820549 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 5 |
Hb_005511_130 |
0.1101120972 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 6 |
Hb_004158_050 |
0.1118452201 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 7 |
Hb_005357_110 |
0.1129193775 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 8 |
Hb_001328_050 |
0.1136061554 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 9 |
Hb_000001_090 |
0.1141633557 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 10 |
Hb_003849_160 |
0.1194166354 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 11 |
Hb_000915_130 |
0.1197920675 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 12 |
Hb_004730_020 |
0.1203607879 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 13 |
Hb_003687_080 |
0.1209178652 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 14 |
Hb_000032_410 |
0.1217996979 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 15 |
Hb_002498_240 |
0.1230634653 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_008494_030 |
0.1251168322 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_002026_020 |
0.1268364745 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 18 |
Hb_002026_090 |
0.1294977037 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 19 |
Hb_000984_220 |
0.1319947362 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 20 |
Hb_164010_050 |
0.1322849422 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |