| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
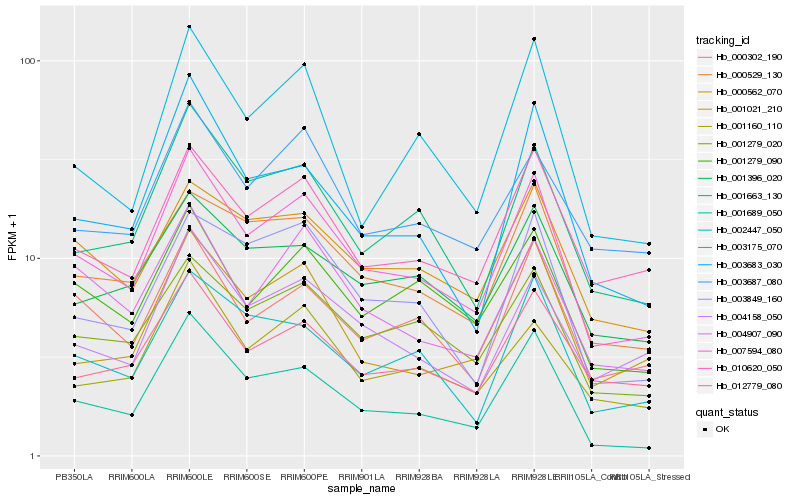
Hb_007594_080 |
0.0 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 2 |
Hb_001279_020 |
0.0873725912 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 3 |
Hb_010620_050 |
0.088430134 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 4 |
Hb_001663_130 |
0.0890095606 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 5 |
Hb_000302_190 |
0.0911171761 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |
| 6 |
Hb_000529_130 |
0.0918763266 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 7 |
Hb_001689_050 |
0.0923040433 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 8 |
Hb_001279_090 |
0.0936728566 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 9 |
Hb_004158_050 |
0.0953132723 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 10 |
Hb_000562_070 |
0.0966098593 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004907_090 |
0.097400989 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_012779_080 |
0.1016653311 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 13 |
Hb_002447_050 |
0.1022140576 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_003683_030 |
0.1031493454 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 15 |
Hb_003175_070 |
0.1036439496 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 16 |
Hb_001160_110 |
0.1040047412 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 17 |
Hb_001396_020 |
0.1040923908 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 18 |
Hb_003687_080 |
0.1044963485 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 19 |
Hb_003849_160 |
0.1046651168 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 20 |
Hb_001021_210 |
0.1054234121 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |