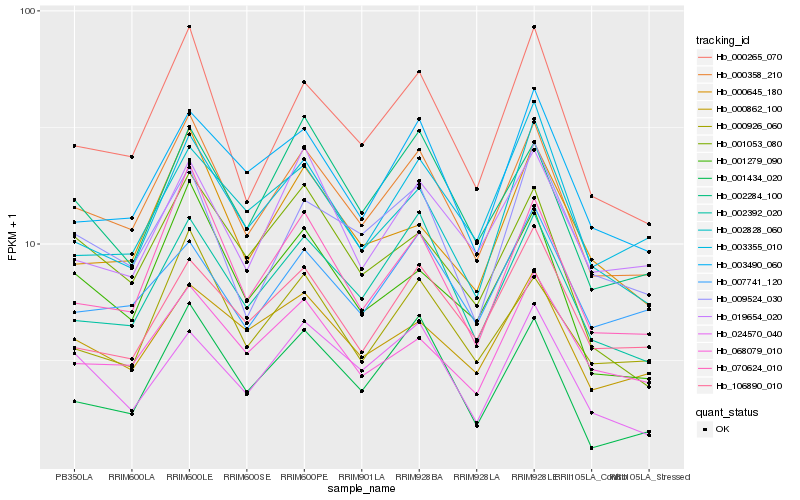
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000358_210 |
0.0 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 2 |
Hb_000265_070 |
0.0604720162 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 3 |
Hb_002392_020 |
0.0715797342 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 4 |
Hb_070624_010 |
0.0726131599 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 5 |
Hb_000926_060 |
0.0830632531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003490_060 |
0.0847997372 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 7 |
Hb_002284_100 |
0.0879800385 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 8 |
Hb_000862_100 |
0.0881590213 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001279_090 |
0.0885140897 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 10 |
Hb_009524_030 |
0.0910435015 |
- |
- |
PREDICTED: uncharacterized protein LOC105650971 [Jatropha curcas] |
| 11 |
Hb_002828_060 |
0.0923672011 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_068079_010 |
0.0929239546 |
- |
- |
- |
| 13 |
Hb_019654_020 |
0.0934780643 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 14 |
Hb_003355_010 |
0.0941927888 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 15 |
Hb_000645_180 |
0.0955278142 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 16 |
Hb_007741_120 |
0.097265551 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001053_080 |
0.0972700215 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 18 |
Hb_001434_020 |
0.0979156504 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 19 |
Hb_106890_010 |
0.0990715943 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_024570_040 |
0.1013966751 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |