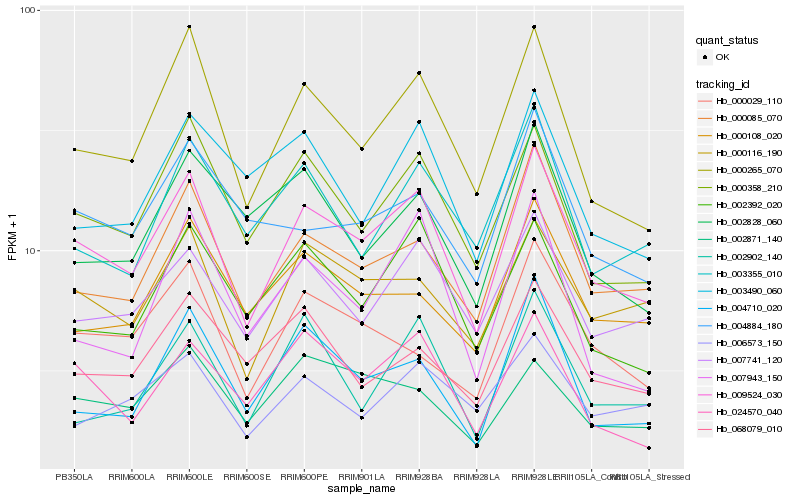
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009524_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650971 [Jatropha curcas] |
| 2 |
Hb_000265_070 |
0.0788751867 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 3 |
Hb_000358_210 |
0.0910435015 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 4 |
Hb_000085_070 |
0.0945118004 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000116_190 |
0.0979665254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007741_120 |
0.0980395072 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_024570_040 |
0.103752435 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 8 |
Hb_002392_020 |
0.1102622263 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 9 |
Hb_000029_110 |
0.1181993123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000108_020 |
0.1192577063 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 11 |
Hb_003355_010 |
0.1202651183 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 12 |
Hb_004710_020 |
0.1212071075 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 13 |
Hb_006573_150 |
0.1222232583 |
- |
- |
PREDICTED: DNA-directed primase/polymerase protein [Jatropha curcas] |
| 14 |
Hb_002871_140 |
0.1227466847 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 15 |
Hb_004884_180 |
0.1229536497 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 16 |
Hb_068079_010 |
0.1234245878 |
- |
- |
- |
| 17 |
Hb_003490_060 |
0.1246226934 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 18 |
Hb_007943_150 |
0.1262473733 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_002828_060 |
0.1270918961 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002902_140 |
0.1272960084 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |