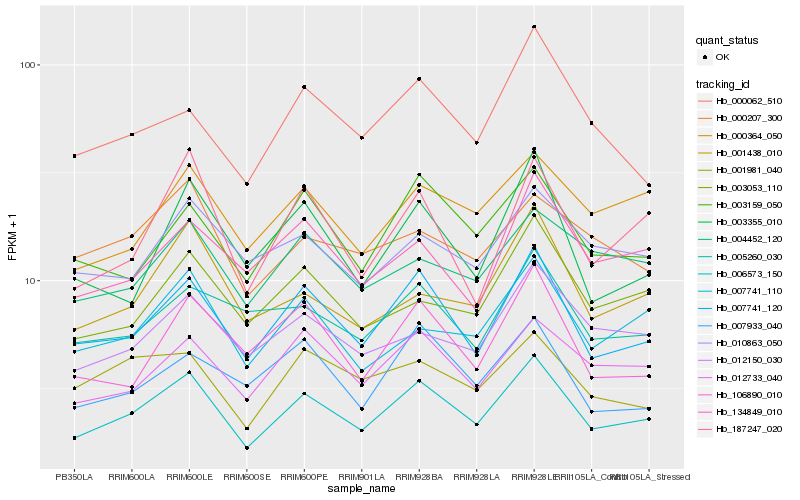
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006573_150 |
0.0 |
- |
- |
PREDICTED: DNA-directed primase/polymerase protein [Jatropha curcas] |
| 2 |
Hb_007741_120 |
0.0579487948 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 3 |
Hb_012733_040 |
0.0862317309 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 4 |
Hb_003159_050 |
0.0923927373 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_007741_110 |
0.0945274058 |
- |
- |
- |
| 6 |
Hb_000364_050 |
0.0966945715 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_012150_030 |
0.0983566388 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_187247_020 |
0.0991502978 |
- |
- |
PREDICTED: uncharacterized protein LOC101220469 [Cucumis sativus] |
| 9 |
Hb_003053_110 |
0.1002747792 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_106890_010 |
0.1026728623 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_134849_010 |
0.1030452647 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 12 |
Hb_004452_120 |
0.1034178989 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 13 |
Hb_010863_050 |
0.1035238003 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 14 |
Hb_001438_010 |
0.1050409968 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005260_030 |
0.1051574104 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 16 |
Hb_001981_040 |
0.1054596569 |
transcription factor |
TF Family: PHD |
PREDICTED: origin of replication complex subunit 1B-like [Jatropha curcas] |
| 17 |
Hb_003355_010 |
0.1054889954 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 18 |
Hb_007933_040 |
0.1056329635 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000062_510 |
0.1080884322 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000207_300 |
0.1087978055 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |