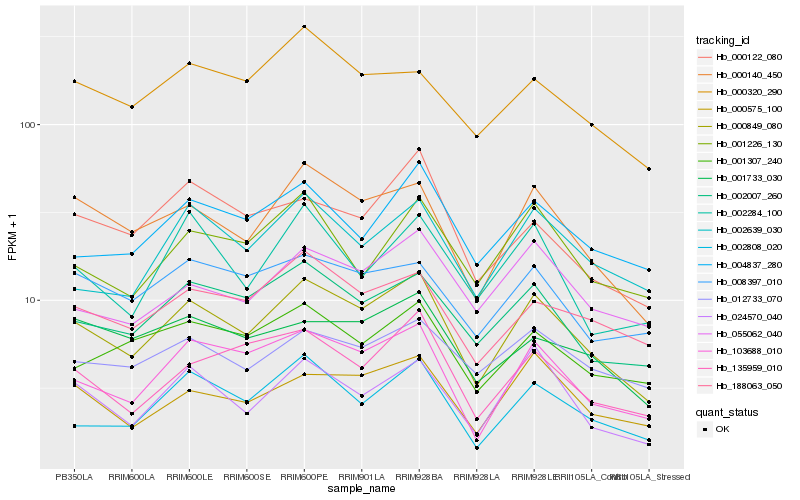
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000849_080 |
0.0 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000140_450 |
0.0763337039 |
- |
- |
PREDICTED: calreticulin-3-like [Populus euphratica] |
| 3 |
Hb_103688_010 |
0.0804906103 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 4 |
Hb_002007_260 |
0.0905091988 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 5 |
Hb_002808_020 |
0.0969393061 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000575_100 |
0.09741863 |
- |
- |
PREDICTED: nephrocystin-3 isoform X2 [Jatropha curcas] |
| 7 |
Hb_024570_040 |
0.0978072705 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 8 |
Hb_188063_050 |
0.1038117577 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 9 |
Hb_135959_010 |
0.1064014494 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 10 |
Hb_055062_040 |
0.107846664 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 11 |
Hb_002639_030 |
0.1082462632 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 12 |
Hb_004837_280 |
0.108864042 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 13 |
Hb_012733_070 |
0.1110646758 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001307_240 |
0.1128364634 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000122_080 |
0.1129981131 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 16 |
Hb_008397_010 |
0.1146142321 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001733_030 |
0.1148887706 |
- |
- |
PREDICTED: uncharacterized protein LOC100261386 isoform X2 [Vitis vinifera] |
| 18 |
Hb_001226_130 |
0.11506289 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 19 |
Hb_002284_100 |
0.1151512445 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 20 |
Hb_000320_290 |
0.1152402035 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |