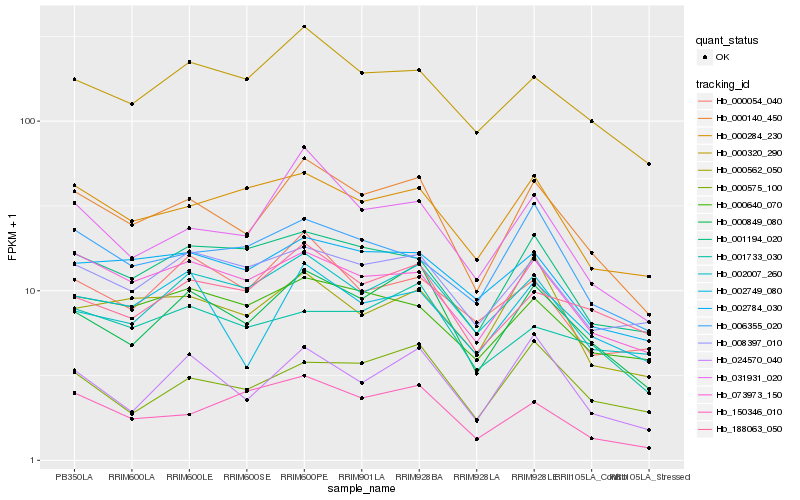
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_450 |
0.0 |
- |
- |
PREDICTED: calreticulin-3-like [Populus euphratica] |
| 2 |
Hb_000849_080 |
0.0763337039 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_031931_020 |
0.0898572881 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 4 |
Hb_073973_150 |
0.0964068583 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000320_290 |
0.0989394526 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 6 |
Hb_001194_020 |
0.1081341504 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 7 |
Hb_000284_230 |
0.1089874696 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 8 |
Hb_002749_080 |
0.1108050915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008397_010 |
0.1114048161 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002784_030 |
0.1117576244 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 11 |
Hb_188063_050 |
0.1133925878 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 12 |
Hb_024570_040 |
0.1133995834 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 13 |
Hb_000640_070 |
0.1141888811 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 14 |
Hb_002007_260 |
0.1160009675 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 15 |
Hb_000562_050 |
0.1172633564 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 16 |
Hb_150346_010 |
0.1199479949 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 17 |
Hb_006355_020 |
0.1206485087 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 18 |
Hb_000054_040 |
0.1223539109 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 19 |
Hb_001733_030 |
0.1225157237 |
- |
- |
PREDICTED: uncharacterized protein LOC100261386 isoform X2 [Vitis vinifera] |
| 20 |
Hb_000575_100 |
0.122639435 |
- |
- |
PREDICTED: nephrocystin-3 isoform X2 [Jatropha curcas] |