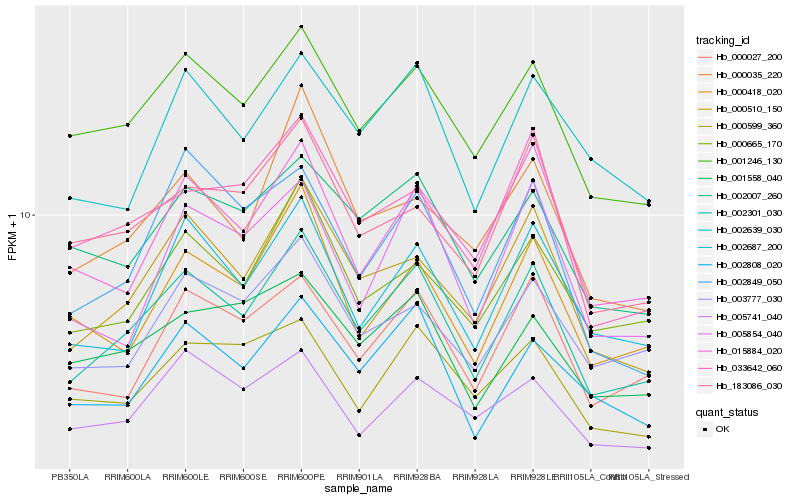
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_030 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 2 |
Hb_000510_150 |
0.0767406098 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001246_130 |
0.0841880083 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 4 |
Hb_002687_200 |
0.0891919787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000027_200 |
0.0896930205 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 6 |
Hb_000665_170 |
0.095112493 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 7 |
Hb_002808_020 |
0.0979974269 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X1 [Jatropha curcas] |
| 8 |
Hb_015884_020 |
0.0995840305 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 9 |
Hb_033642_060 |
0.0996349578 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003777_030 |
0.0999660153 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 11 |
Hb_002007_260 |
0.1000298815 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 12 |
Hb_000418_020 |
0.1000425397 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001558_040 |
0.1020136499 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 14 |
Hb_005854_040 |
0.1033844979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002639_030 |
0.1035276048 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 16 |
Hb_002849_050 |
0.1044676614 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 17 |
Hb_000035_220 |
0.1048231339 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 18 |
Hb_183086_030 |
0.1050402211 |
- |
- |
GTPase-activating protein GYP7 [Gossypium arboreum] |
| 19 |
Hb_005741_040 |
0.107460504 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 20 |
Hb_000599_360 |
0.1084491542 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |