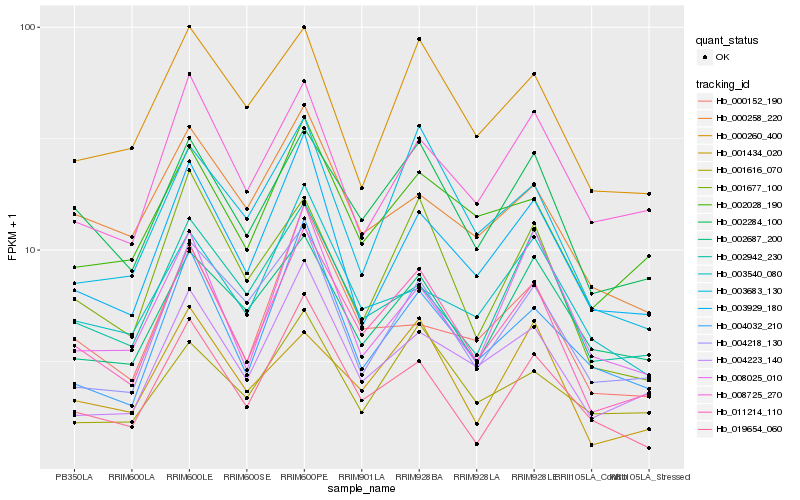
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011214_110 |
0.0 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 2 |
Hb_002284_100 |
0.08832164 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 3 |
Hb_004223_140 |
0.104969794 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 4 |
Hb_001677_100 |
0.105997934 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 5 |
Hb_003929_180 |
0.106656984 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 6 |
Hb_019654_060 |
0.1068570225 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004032_210 |
0.1104444669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002028_190 |
0.1121171066 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7-like [Jatropha curcas] |
| 9 |
Hb_000152_190 |
0.1160629055 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |
| 10 |
Hb_001434_020 |
0.11880677 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 11 |
Hb_003683_130 |
0.1193133797 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002942_230 |
0.1200461673 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 13 |
Hb_000258_220 |
0.1245218592 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000260_400 |
0.1252647733 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 15 |
Hb_008725_270 |
0.125932286 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 16 |
Hb_003540_080 |
0.1262331209 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_004218_130 |
0.1281427258 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 18 |
Hb_008025_010 |
0.1285925444 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |
| 19 |
Hb_001616_070 |
0.1286352905 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 20 |
Hb_002687_200 |
0.1287928025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |