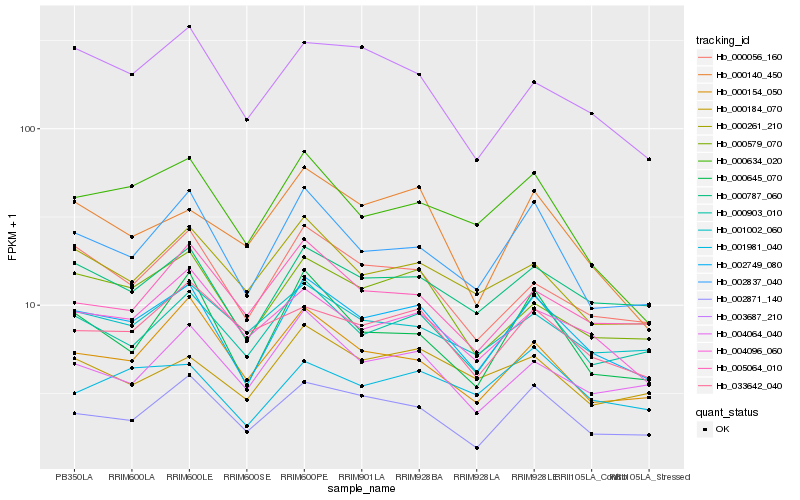
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002749_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002837_040 |
0.081639155 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 3 |
Hb_000787_060 |
0.0825681371 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000903_010 |
0.0829695741 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 5 |
Hb_002871_140 |
0.0862947037 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 6 |
Hb_000579_070 |
0.0877528908 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 7 |
Hb_000056_160 |
0.0922751984 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP23 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000645_070 |
0.0938451826 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_004096_060 |
0.0992971997 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 10 |
Hb_000261_210 |
0.1011894098 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 11 |
Hb_000634_020 |
0.1018401387 |
- |
- |
hypothetical protein JCGZ_11888 [Jatropha curcas] |
| 12 |
Hb_000154_050 |
0.1033924343 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 13 |
Hb_004064_040 |
0.1045264117 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 14 |
Hb_003687_210 |
0.107219217 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 15 |
Hb_000184_070 |
0.1103718457 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000140_450 |
0.1108050915 |
- |
- |
PREDICTED: calreticulin-3-like [Populus euphratica] |
| 17 |
Hb_001981_040 |
0.1115601823 |
transcription factor |
TF Family: PHD |
PREDICTED: origin of replication complex subunit 1B-like [Jatropha curcas] |
| 18 |
Hb_001002_060 |
0.1116692215 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 19 |
Hb_005064_010 |
0.1126684357 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 20 |
Hb_033642_040 |
0.1127592586 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |