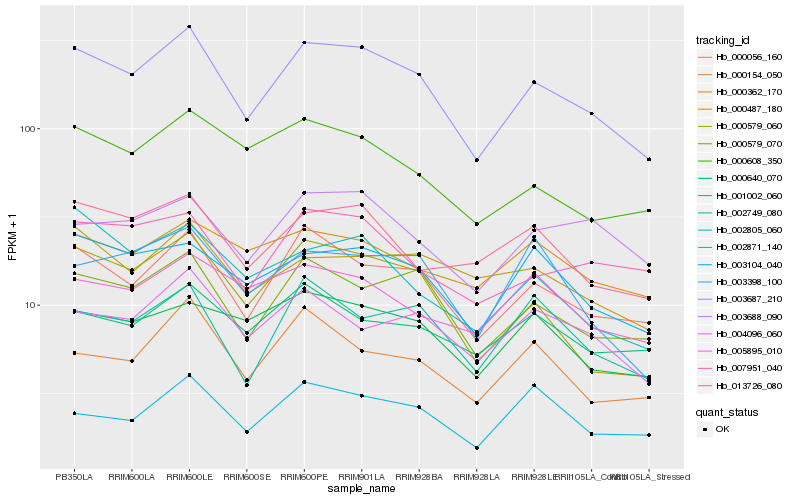
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003687_210 |
0.0 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 2 |
Hb_000056_160 |
0.0680730347 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP23 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000579_070 |
0.0777452869 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 4 |
Hb_000579_060 |
0.0892508313 |
- |
- |
PREDICTED: uncharacterized protein LOC105633849 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003104_040 |
0.092964337 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 6 |
Hb_013726_080 |
0.1013347289 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 7 |
Hb_004096_060 |
0.1030330027 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 8 |
Hb_000608_350 |
0.1037200653 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 9 |
Hb_000640_070 |
0.1044201154 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 10 |
Hb_002749_080 |
0.107219217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007951_040 |
0.1072264511 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 12 |
Hb_003688_090 |
0.1097414761 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000154_050 |
0.1098511932 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 14 |
Hb_001002_060 |
0.111411675 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 15 |
Hb_002805_060 |
0.1115037424 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 16 |
Hb_000487_180 |
0.1127587691 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002871_140 |
0.1132416965 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 18 |
Hb_005895_010 |
0.1138003788 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003398_100 |
0.1144871101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000362_170 |
0.1152534377 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |