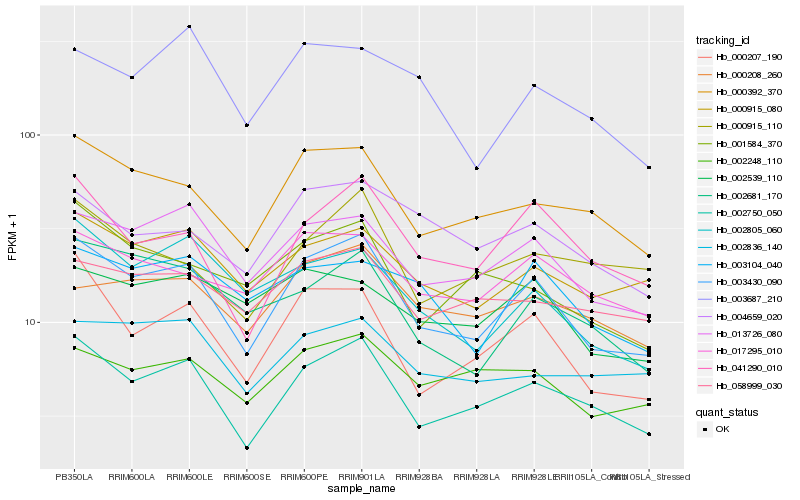
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003430_090 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_002750_050 |
0.0683343908 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 3 |
Hb_013726_080 |
0.0883145767 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 4 |
Hb_000392_370 |
0.0921572455 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 5 |
Hb_000207_190 |
0.0935371015 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 6 |
Hb_041290_010 |
0.1035137486 |
- |
- |
thioredoxin family protein [Populus trichocarpa] |
| 7 |
Hb_000915_080 |
0.1076391375 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 8 |
Hb_002805_060 |
0.1108070518 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 9 |
Hb_017295_010 |
0.1123388044 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 10 |
Hb_002681_170 |
0.1135647087 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 11 |
Hb_002248_110 |
0.1178854509 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 12 |
Hb_001584_370 |
0.1224209313 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 13 |
Hb_004659_020 |
0.1227722562 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 14 |
Hb_000208_260 |
0.1229747148 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 15 |
Hb_002836_140 |
0.123509416 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 16 |
Hb_003104_040 |
0.1241848702 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 17 |
Hb_058999_030 |
0.1247576527 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_003687_210 |
0.1257140969 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 19 |
Hb_002539_110 |
0.1268179161 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 20 |
Hb_000915_110 |
0.1286256336 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |