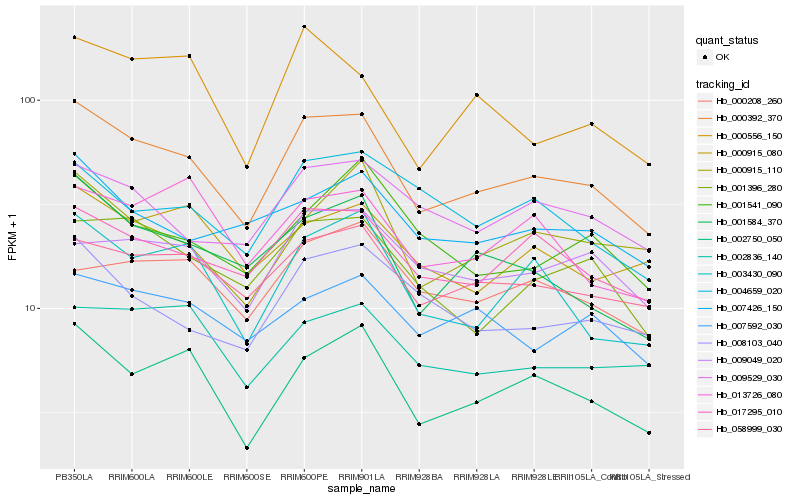
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_370 |
0.0 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 2 |
Hb_017295_010 |
0.0812293703 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 3 |
Hb_002750_050 |
0.0822935118 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 4 |
Hb_058999_030 |
0.0856637763 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_001584_370 |
0.0903317122 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 6 |
Hb_003430_090 |
0.0921572455 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_001396_280 |
0.095479945 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 8 |
Hb_000915_110 |
0.1013378427 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 9 |
Hb_008103_040 |
0.1020821721 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_009529_030 |
0.1023482702 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 11 |
Hb_007592_030 |
0.1033744285 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_002836_140 |
0.1036551813 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 13 |
Hb_007426_150 |
0.1046608455 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 14 |
Hb_009049_020 |
0.1050164605 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 15 |
Hb_000556_150 |
0.1057331479 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 16 |
Hb_000915_080 |
0.1065299238 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 17 |
Hb_001541_090 |
0.108866099 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004659_020 |
0.1099089687 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 19 |
Hb_000208_260 |
0.1100736971 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_013726_080 |
0.1101651799 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |