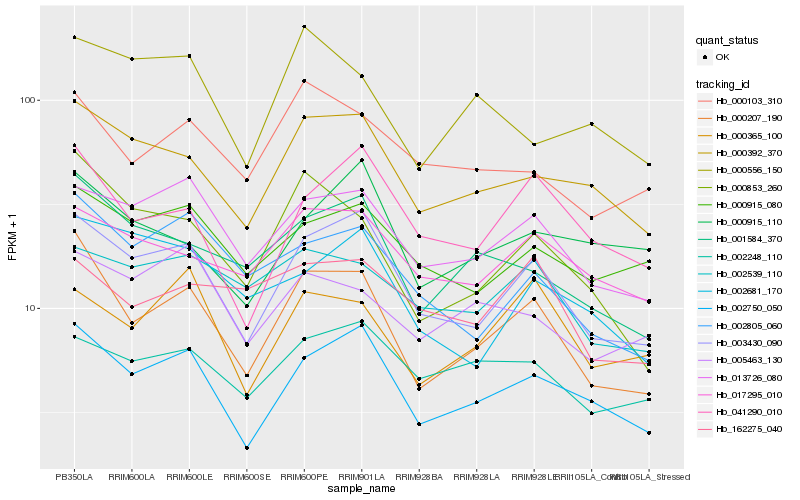
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000207_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 2 |
Hb_003430_090 |
0.0935371015 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_002750_050 |
0.1015708112 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 4 |
Hb_000853_260 |
0.1017872711 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 5 |
Hb_001584_370 |
0.1188048083 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 6 |
Hb_000392_370 |
0.120575926 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 7 |
Hb_002805_060 |
0.1297149581 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 8 |
Hb_013726_080 |
0.1310819263 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 9 |
Hb_041290_010 |
0.1392932426 |
- |
- |
thioredoxin family protein [Populus trichocarpa] |
| 10 |
Hb_000103_310 |
0.1422701354 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 11 |
Hb_017295_010 |
0.1491133791 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 12 |
Hb_000365_100 |
0.1508956032 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000915_080 |
0.1512641023 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 14 |
Hb_002539_110 |
0.1542579327 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 15 |
Hb_005463_130 |
0.1559034439 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 16 |
Hb_002681_170 |
0.1578114573 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 17 |
Hb_002248_110 |
0.1597162302 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 18 |
Hb_000556_150 |
0.1603382619 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 19 |
Hb_162275_040 |
0.1649488397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000915_110 |
0.1652321366 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |