| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
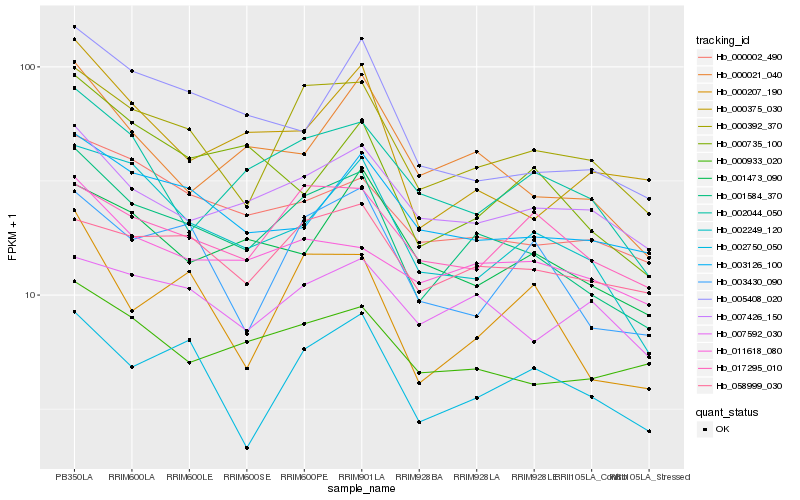
Hb_001584_370 |
0.0 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 2 |
Hb_000392_370 |
0.0903317122 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 3 |
Hb_000021_040 |
0.1059528 |
- |
- |
PREDICTED: villin-3-like isoform X1 [Populus euphratica] |
| 4 |
Hb_000002_490 |
0.1063765765 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 5 |
Hb_002249_120 |
0.1075809214 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 6 |
Hb_007426_150 |
0.1120026716 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 7 |
Hb_000933_020 |
0.1150134464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_058999_030 |
0.1164079575 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_005408_020 |
0.1176807188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003126_100 |
0.1181945455 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 11 |
Hb_011618_080 |
0.1185601621 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 12 |
Hb_002044_050 |
0.1186343355 |
- |
- |
- |
| 13 |
Hb_000207_190 |
0.1188048083 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 14 |
Hb_002750_050 |
0.1220904589 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 15 |
Hb_003430_090 |
0.1224209313 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000735_100 |
0.1224359029 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 17 |
Hb_007592_030 |
0.1233857576 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 18 |
Hb_017295_010 |
0.1240315117 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 19 |
Hb_001473_090 |
0.1242352932 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 20 |
Hb_000375_030 |
0.1243137079 |
- |
- |
- |