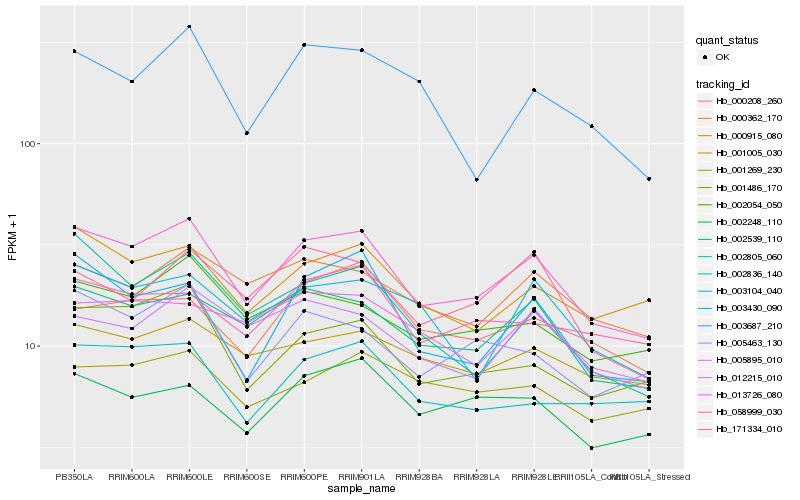
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013726_080 |
0.0 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 2 |
Hb_002539_110 |
0.0704555149 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 3 |
Hb_001486_170 |
0.0830608218 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 4 |
Hb_000915_080 |
0.085204768 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 5 |
Hb_005463_130 |
0.0866608475 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 6 |
Hb_002054_050 |
0.0868714882 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 7 |
Hb_003430_090 |
0.0883145767 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_005895_010 |
0.0912477756 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000362_170 |
0.0928344501 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 10 |
Hb_012215_010 |
0.0929179847 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 11 |
Hb_003104_040 |
0.0936085588 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 12 |
Hb_002836_140 |
0.0936756063 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 13 |
Hb_002248_110 |
0.0937516384 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 14 |
Hb_058999_030 |
0.0952944069 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_001005_030 |
0.0957704534 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 16 |
Hb_171334_010 |
0.09618168 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 17 |
Hb_002805_060 |
0.0962990448 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 18 |
Hb_003687_210 |
0.1013347289 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 19 |
Hb_000208_260 |
0.1025147497 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_001269_230 |
0.102775411 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |