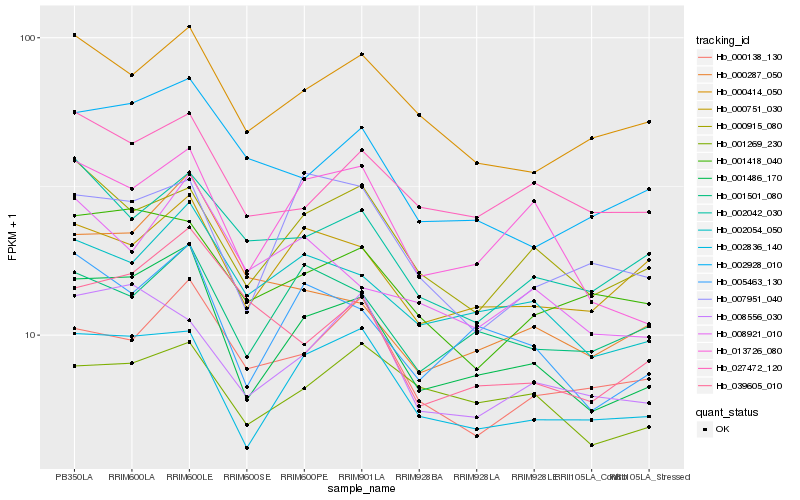
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_170 |
0.0 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 2 |
Hb_005463_130 |
0.0696645374 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 3 |
Hb_002836_140 |
0.0727420733 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 4 |
Hb_013726_080 |
0.0830608218 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 5 |
Hb_002054_050 |
0.0839094888 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 6 |
Hb_000915_080 |
0.0929184754 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 7 |
Hb_008556_030 |
0.0949596497 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001501_080 |
0.0960818683 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000751_030 |
0.0961489468 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 10 |
Hb_002928_010 |
0.0964567686 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 11 |
Hb_000414_050 |
0.0966215612 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 12 |
Hb_027472_120 |
0.0968775717 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_001418_040 |
0.1010044988 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 14 |
Hb_000138_130 |
0.1066638936 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |
| 15 |
Hb_007951_040 |
0.106932345 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 16 |
Hb_008921_010 |
0.1077671284 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 17 |
Hb_002042_030 |
0.10811784 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 18 |
Hb_000287_050 |
0.1082776058 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 19 |
Hb_001269_230 |
0.1090228937 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 20 |
Hb_039605_010 |
0.110666631 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |