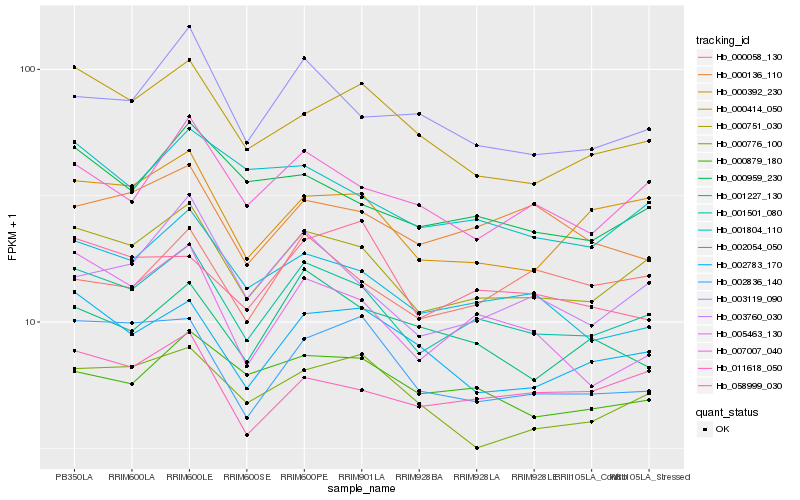
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001501_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000751_030 |
0.0329206946 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 3 |
Hb_002054_050 |
0.0716497801 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 4 |
Hb_005463_130 |
0.0765149871 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 5 |
Hb_000392_230 |
0.0789878855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007007_040 |
0.0790972384 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 7 |
Hb_003760_030 |
0.0801604866 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 8 |
Hb_003119_090 |
0.0806522941 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 9 |
Hb_000879_180 |
0.0814451477 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 10 |
Hb_011618_050 |
0.0820961987 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 11 |
Hb_000959_230 |
0.0826694836 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 12 |
Hb_002836_140 |
0.0844346675 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 13 |
Hb_002783_170 |
0.0854790931 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 14 |
Hb_000136_110 |
0.0865270841 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 15 |
Hb_000414_050 |
0.087357014 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 16 |
Hb_000058_130 |
0.08738093 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 17 |
Hb_001227_130 |
0.0880731657 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 18 |
Hb_001804_110 |
0.0885551068 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 19 |
Hb_058999_030 |
0.0893637841 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_000776_100 |
0.0897645746 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |