| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
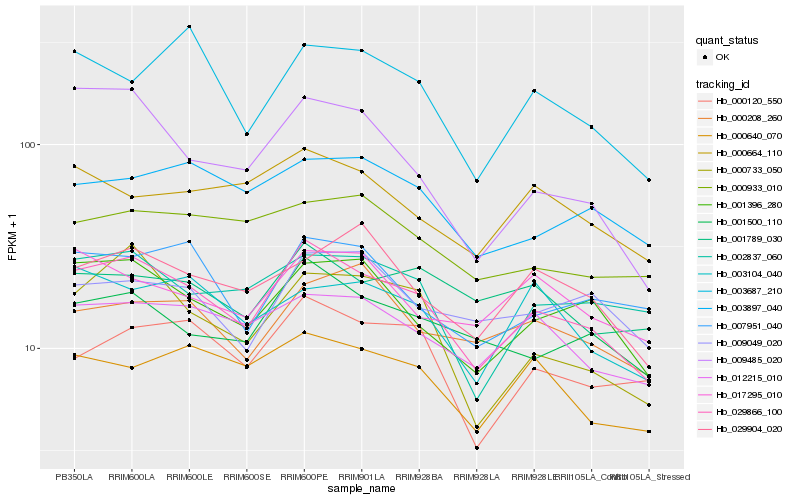
Hb_029866_100 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 2 |
Hb_001396_280 |
0.0840451322 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 3 |
Hb_001500_110 |
0.0957073461 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_012215_010 |
0.0976618455 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 5 |
Hb_000733_050 |
0.1038073985 |
- |
- |
PREDICTED: uncharacterized protein LOC105646910 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002837_060 |
0.1065532717 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 7 |
Hb_000120_550 |
0.1073769295 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 8 |
Hb_000640_070 |
0.1085671445 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 9 |
Hb_007951_040 |
0.1099498213 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 10 |
Hb_003897_040 |
0.1109649356 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 11 |
Hb_009485_020 |
0.1113907573 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 12 |
Hb_017295_010 |
0.1125087246 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 13 |
Hb_000664_110 |
0.1132823367 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 14 |
Hb_009049_020 |
0.1137288959 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 15 |
Hb_029904_020 |
0.114403126 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 16 |
Hb_003104_040 |
0.1150092155 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 17 |
Hb_003687_210 |
0.1153326642 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 18 |
Hb_000933_010 |
0.1154983144 |
- |
- |
- |
| 19 |
Hb_001789_030 |
0.1164647198 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 20 |
Hb_000208_260 |
0.1168094029 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |