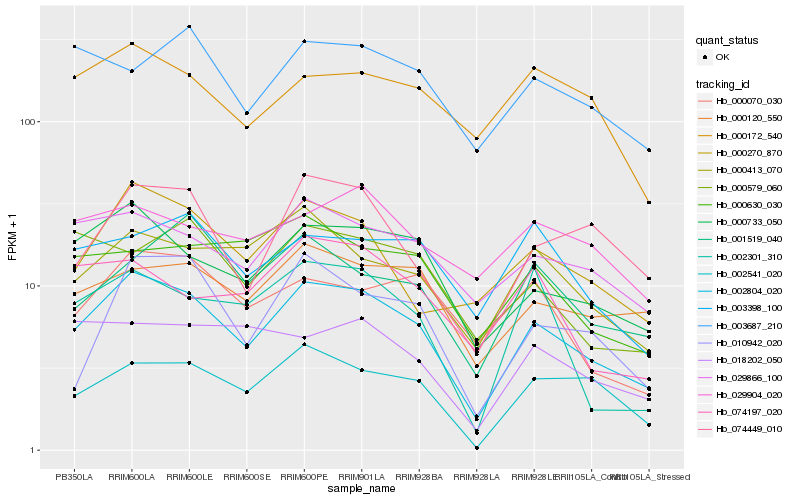
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002804_020 |
0.0 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_000733_050 |
0.1153729892 |
- |
- |
PREDICTED: uncharacterized protein LOC105646910 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002541_020 |
0.1299410762 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 4 |
Hb_029866_100 |
0.1323834242 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 5 |
Hb_000270_870 |
0.1332427248 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 6 |
Hb_074449_010 |
0.1384260096 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 7 |
Hb_000413_070 |
0.1389490905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000120_550 |
0.1397620995 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 9 |
Hb_010942_020 |
0.15027163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000070_030 |
0.1523866475 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 11 |
Hb_001519_040 |
0.156407202 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 12 |
Hb_000172_540 |
0.1567741767 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |
| 13 |
Hb_003398_100 |
0.1582551646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003687_210 |
0.160294803 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 15 |
Hb_002301_310 |
0.1605267993 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_018202_050 |
0.1620565459 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A [Jatropha curcas] |
| 17 |
Hb_000579_060 |
0.1623608415 |
- |
- |
PREDICTED: uncharacterized protein LOC105633849 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000630_030 |
0.1623638284 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 19 |
Hb_074197_020 |
0.1624033609 |
- |
- |
PREDICTED: uncharacterized protein LOC105645948 [Jatropha curcas] |
| 20 |
Hb_029904_020 |
0.1626801154 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |