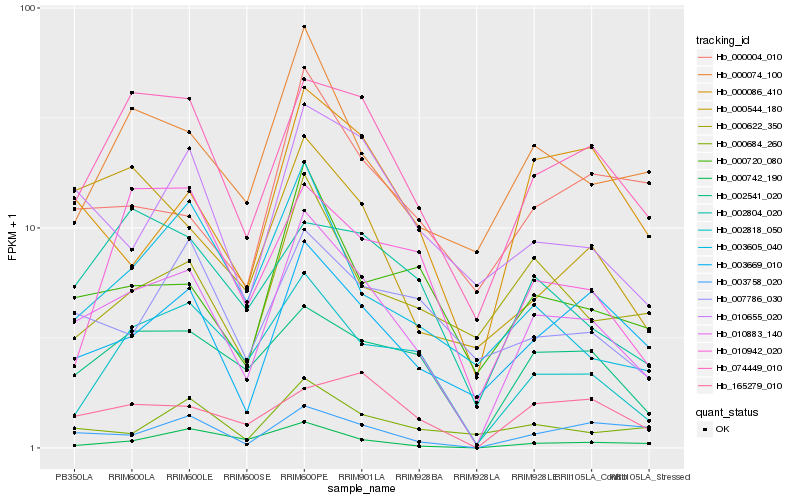
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010883_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_074449_010 |
0.1424153941 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 3 |
Hb_002541_020 |
0.1539641013 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 4 |
Hb_003669_010 |
0.1672716393 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic [Populus euphratica] |
| 5 |
Hb_002818_050 |
0.1766766754 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 6 |
Hb_010655_020 |
0.1785264048 |
- |
- |
hypothetical protein CICLE_v10032410mg [Citrus clementina] |
| 7 |
Hb_000074_100 |
0.1797011737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000720_080 |
0.1808770191 |
- |
- |
PREDICTED: bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 [Jatropha curcas] |
| 9 |
Hb_003758_020 |
0.1825900362 |
- |
- |
UGTPg34 [Panax ginseng] |
| 10 |
Hb_002804_020 |
0.186229081 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 11 |
Hb_165279_010 |
0.1866045234 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 12 |
Hb_000086_410 |
0.1886171054 |
- |
- |
hypothetical protein CISIN_1g0263261mg [Citrus sinensis] |
| 13 |
Hb_000544_180 |
0.1896011019 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X1 [Jatropha curcas] |
| 14 |
Hb_000622_350 |
0.1947523297 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 15 |
Hb_003605_040 |
0.1977033722 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007786_030 |
0.1991285923 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 17 |
Hb_000684_260 |
0.2001448984 |
- |
- |
PREDICTED: probable DNA helicase MCM8 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000004_010 |
0.2027638413 |
- |
- |
hypothetical protein Csa_6G120400 [Cucumis sativus] |
| 19 |
Hb_000742_190 |
0.2043599288 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_010942_020 |
0.2048693257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |