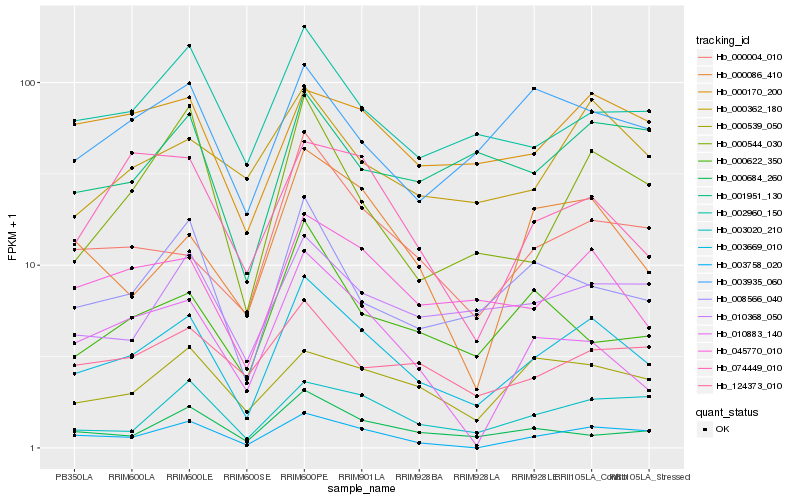
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003669_010 |
0.0 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic [Populus euphratica] |
| 2 |
Hb_045770_010 |
0.1285978241 |
- |
- |
ER lumen protein retaining receptor [Populus trichocarpa] |
| 3 |
Hb_003758_020 |
0.1404336529 |
- |
- |
UGTPg34 [Panax ginseng] |
| 4 |
Hb_000004_010 |
0.1462431785 |
- |
- |
hypothetical protein Csa_6G120400 [Cucumis sativus] |
| 5 |
Hb_008566_040 |
0.1483259407 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002960_150 |
0.1521686417 |
- |
- |
hypothetical protein CISIN_1g029215mg [Citrus sinensis] |
| 7 |
Hb_003020_210 |
0.1543661058 |
- |
- |
Structural maintenance of chromosomes 2-2 -like protein [Gossypium arboreum] |
| 8 |
Hb_010368_050 |
0.1564137227 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 9 |
Hb_000544_030 |
0.1572047162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000086_410 |
0.158912202 |
- |
- |
hypothetical protein CISIN_1g0263261mg [Citrus sinensis] |
| 11 |
Hb_000362_180 |
0.1589507011 |
- |
- |
Pdap1 [Gossypium arboreum] |
| 12 |
Hb_000539_050 |
0.1623657851 |
- |
- |
bile acid:sodium symporter, putative [Ricinus communis] |
| 13 |
Hb_000684_260 |
0.1626704235 |
- |
- |
PREDICTED: probable DNA helicase MCM8 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000170_200 |
0.1628141903 |
- |
- |
PREDICTED: protein KRTCAP2 homolog [Jatropha curcas] |
| 15 |
Hb_124373_010 |
0.1658400113 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 16 |
Hb_074449_010 |
0.166307852 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 17 |
Hb_003935_060 |
0.1671696115 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 18 |
Hb_010883_140 |
0.1672716393 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000622_350 |
0.1703927146 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 20 |
Hb_001951_130 |
0.1721606621 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |