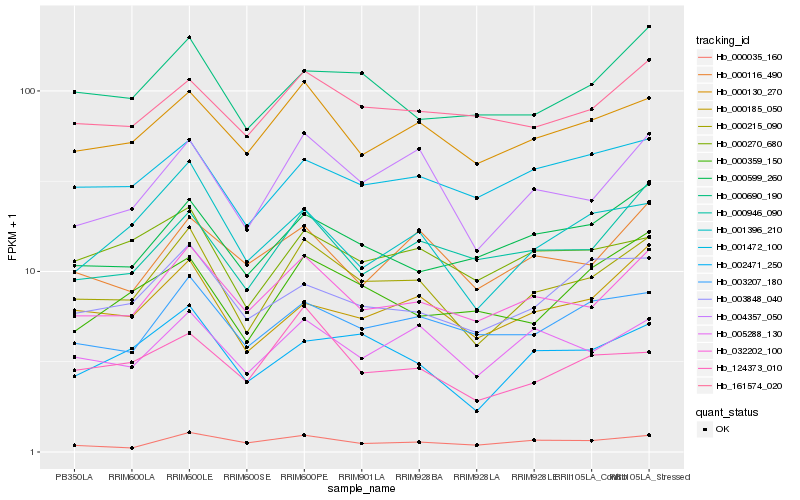
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_090 |
0.0 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_032202_100 |
0.084285931 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 3 |
Hb_004357_050 |
0.0843857819 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 4 |
Hb_000130_270 |
0.0940692521 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002471_250 |
0.0989036392 |
- |
- |
Tapt1/CMV receptor, putative isoform 2 [Theobroma cacao] |
| 6 |
Hb_005288_130 |
0.1028567737 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_003207_180 |
0.1055519719 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000185_050 |
0.1066076801 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 9 |
Hb_000690_190 |
0.1087922841 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 10 |
Hb_001396_210 |
0.1096592834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001472_100 |
0.1102449659 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 12 |
Hb_000599_260 |
0.1110937157 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 13 |
Hb_000270_680 |
0.1122376336 |
- |
- |
hypothetical protein POPTR_0015s08420g [Populus trichocarpa] |
| 14 |
Hb_003848_040 |
0.1127808397 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 15 |
Hb_000946_090 |
0.1127884528 |
- |
- |
PREDICTED: cyclin-C1-2-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_161574_020 |
0.1129324323 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 17 |
Hb_000035_160 |
0.1147833101 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 18 |
Hb_124373_010 |
0.1179266217 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 19 |
Hb_000359_150 |
0.1188496681 |
- |
- |
PREDICTED: protein Mpv17-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000116_490 |
0.1192764659 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |